













| General Information: |
|
| Name(s) found: |
MAP22_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 439 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA]
|
| Biological Process: |
protein processing
[TAS]
|
| Molecular Function: |
metalloexopeptidase activity
[IEA]
aminopeptidase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASESPDVAV VAPVVENGGA ESSNGKEEQL ESELSKKLEI AEDGQEENDG EEGSKAETST 60
61 KKKKKKNKSK KKKELPQQTD PPSIPVVELF PSGEFPEGEI QEYKDDNLWR TTSEEKRELE 120
121 RFEKPIYNSV RRAAEVHRQV RKYVRSIVKP GMLMTDICET LENTVRKLIS ENGLQAGIAF 180
181 PTGCSLNWVA AHWTPNSGDK TVLQYDDVMK LDFGTHIDGH IIDCAFTVAF NPMFDPLLAA 240
241 SREATYTGIK EAGIDVRLCD IGAAIQEVME SYEVEINGKV FQVKSIRNLN GHSIGPYQIH 300
301 AGKSVPIVKG GEQTKMEEGE FYAIETFGST GKGYVREDLE CSHYMKNFDA GHVPLRLPRA 360
361 KQLLATINKN FSTLAFCRRY LDRIGETKYL MALKNLCDSG IVQPYPPLCD VKGSYVSQFE 420
421 HTILLRPTCK EVLSKGDDY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
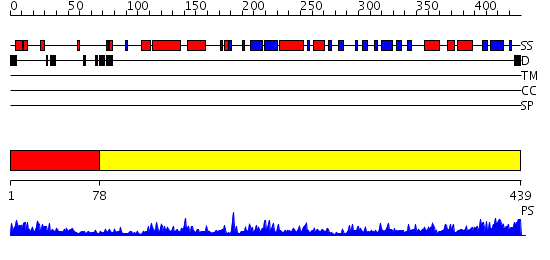
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..77] | 58.0 | AMINOPEPTIDASE P FROM E. COLI WITH THE INHIBITOR PRO-LEU | |
| 2 | View Details | [78..439] | 102.0 | Methionine aminopeptidase, insert domain; Methionine aminopeptidase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)