













| General Information: |
|
| Name(s) found: |
gi|18414189
[NCBI NR]
gi|16973453 [NCBI NR] gi|16973451 [NCBI NR] |
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 869 amino acids |
Gene Ontology: |
|
| Cellular Component: |
phragmoplast
[IDA]
|
| Biological Process: |
microtubule-based movement
[IEA]
|
| Molecular Function: |
ATP binding
[IEA]
microtubule motor activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAPTPSSSRS NQTQYTLIRT PQTKQRLNFH SKTPNPDGSK DPSPPEHPVE VIGRIRDYPD 60
61 RKEKSPSILQ VNTDNQTVRV RADVGYRDFT LDGVSFSEQE GLEEFYKKFI EERIKGVKVG 120
121 NKCTIMMYGP TGAGKSHTMF GCGKEPGIVY RSLRDILGDS DQDGVTFVQV TVLEVYNEEI 180
181 YDLLSTNSSN NLGIGWPKGA STKVRLEVMG KKAKNASFIS GTEAGKISKE IVKVEKRRIV 240
241 KSTLCNERSS RSHCIIILDV PTVGGRLMLV DMAGSENIDQ AGQTGFEAKM QTAKINQGNI 300
301 ALKRVVESIA NGDSHVPFRD SKLTMLLQDS FEDDKSKILM ILCASPDPKE MHKTLCTLEY 360
361 GAKAKCIVRG SHTPNKDKYG GDESASAVIL GSRIAAMDEF IIKLQSEKKQ KEKERNEAQK 420
421 QLKKKEEEVA ALRSLLTQRE ACATNEEEIK EKVNERTQLL KSELDKKLEE CRRMAEEFVE 480
481 MERRRMEERI VQQQEELEMM RRRLEEIEVE FRRSNGGSVD ETSGFAKRLR SLYSDDDPGM 540
541 VKSMDLDMGD PEPVKQVWGA VSHQSSNTIS SNFTNLLQPK PSENMLTQMY PDRVCLSTVF 600
601 EEEEVEEEEE KVIVEDKSIC LITTPMPSLN SEGLGKENCF NGADDKESAS SRRLRIQNIF 660
661 TLCGNQRELS QHSGQEEDQA NIASPDKKDN QFFSITNKAE ALAVEEAKEN NISVDQRENG 720
721 QLDIYVKWET AADNPRKLIT TLRVTKDATL ADLRKLIEIY LGSDNQAFTF LKLGEPCGAQ 780
781 VAKEKESTVQ ATSLPLCNGH AYLATLRPGK SSQHKSLQPA SPLPLNPIEN MMEVTPISKV 840
841 TPNHQVDEFS SPNLVAHLSS TPFITLRRH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
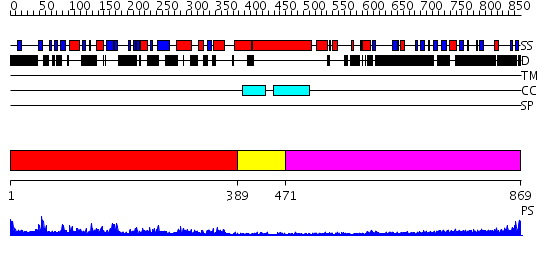
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..388] | 72.154902 | Kinesin | |
| 2 | View Details | [389..470] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [471..869] | 10.221849 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)