













| General Information: |
|
| Name(s) found: |
CLSY2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1261 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: |
nucleic acid binding
[IEA]
ATP binding [IEA][ISS] DNA binding [IEA][ISS] helicase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKKRGFYNLK HPFDPCPFEF FCSGTWKPVE YMRIEDGMMT IRLLENGYVL EDIRPFQRLR 60
61 LRSRKAALSD CICFLRPDID VCVLYRIHED DLEPVWVDAR IVSIERKPHE SECSCKINVR 120
121 IYIDQGCIGS EKQRINRDSV VIGLNQISIL QKFYKEQSTD QFYRWRFSED CTSLMKTRLS 180
181 LGKFLPDLSW LTVTSTLKSI VFQIRTVQTK MVYQIVTDEE GSSSTLSSMN ITLEDGVSLS 240
241 KVVKFNPADI LDDSQDLEIK QETDYYQEED EVVELRRSKR RNVRPDIYTG CDYEPDTIDG 300
301 WVRMMPYQFG KCAVNVESDE DEDDNNEDGD TNDDLYIPLS RLFIKKKKTN SREAKPKSRK 360
361 GEIVVIDKRR VHGFGRKERK SELSVIPFTP VFEPIPLEQF GLNANSFGGG GSFSRSQYFD 420
421 ETEKYRSKGM KYGKKMTEME EMMEADLCWK GPNQVKSFQK RTSRSSRSVA PKTEDSDEPR 480
481 VYKKVTLSAG AYNKLIDTYM NNIESTIAAK DEPTSVVDQW EELKKTNFAF KLHGDMEKNL 540
541 SEDGEGETSE NEMLWREMEL CLASSYILDD NEVRVDNEAF EKARSGCEHD YRLEEEIGMC 600
601 CRLCGHVGSE IKDVSAPFAE HKKWTIETKH IEEDDIKTKL SHKEAQTKDF SMISDSSEML 660
661 AAEESDNVWA LIPKLKRKLH VHQRRAFEFL WRNVAGSVEP SLMDPTSGNI GGCVISHSPG 720
721 AGKTFLIIAF LTSYLKLFPG KRPLVLAPKT TLYTWYKEFI KWEIPVPVHL IHGRRTYCTF 780
781 KQNKTVQFNG VPKPSRDVMH VLDCLEKIQK WHAHPSVLVM GYTSFTTLMR EDSKFAHRKY 840
841 MAKVLRESPG LLVLDEGHNP RSTKSRLRKA LMKVGTDLRI LLSGTLFQNN FCEYFNTLCL 900
901 ARPKFIHEVL MELDQKFKTN HGVNKAPHLL ENRARKLFLD IIAKKIDASV GDERLQGLNM 960
961 LKNMTNGFID NYEGSGSGSG DALPGLQIYT LVMNSTDIQH KILTKLQDVI KTYFGYPLEV1020
1021 ELQITLAAIH PWLVTSSNCC TKFFNPQELS EIGKLKHDAK KGSKVMFVLN LIFRVVKREK1080
1081 ILIFCHNIAP IRMFTELFEN IFRWQRGREI LTLTGDLELF ERGRVIDKFE EPGNPSRVLL1140
1141 ASITACAEGI SLTAASRVIM LDSEWNPSKT KQAIARAFRP GQQKVVYVYQ LLSRGTLEED1200
1201 KYRRTTWKEW VSCMIFSEEF VADPSLWQAE KIEDDILREI VGEDKVKSFH MIMKNEKAST1260
1261 G |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
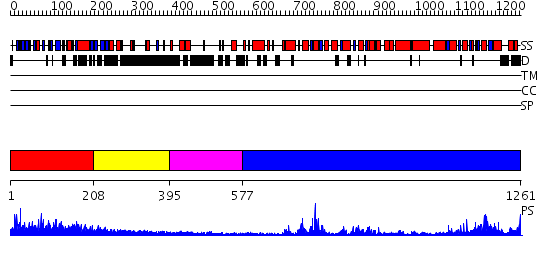
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..207] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [208..394] | 2.360996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [395..576] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [577..1261] | 75.0 | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)