













| LEGEND: |
|||
| = Same process, or one is unknown. | = Same branch, distance 4. | ||
| = Same branch, distance 1. | = Same branch, distance 5. | ||
| = Same branch, distance 2. | = Same branch, distance 6 or more. | ||
| = Same branch, distance 3. | = Not in same branch of GO. | ||
| Size | Notes | Members | |
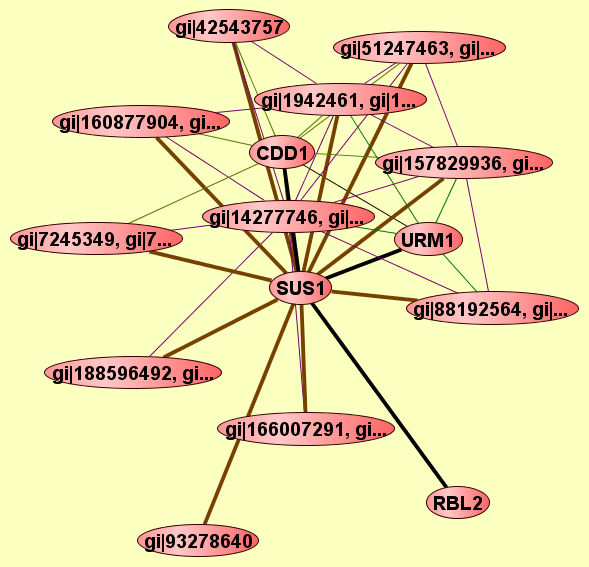
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 1IGNA and 3FWBC with confidence: 0.987927466143 | SUS1 gi|1942461, gi|1... |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 1R5TA and 3FWBC with confidence: 0.989995527752 | CDD1 SUS1 |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 1D1QA and 3FWBC with confidence: 0.907476513329 | SUS1 gi|7245349, gi|7... |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 1I12A and 3FWBC with confidence: 0.990147465316 | SUS1 gi|14277743, gi|... |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 1AKYA and 3FWBC with confidence: 0.982912352423 | SUS1 gi|157829936, gi... |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 2Z5BA and 3FWBC with confidence: 0.932267969741 | SUS1 gi|166007291, gi... |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 3CZ6A and 3FWBC with confidence: 0.927376339539 | SUS1 gi|188596492, gi... |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 1SZ9A and 3FWBC with confidence: 0.947859062345 | SUS1 gi|51247462, gi|... |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 2QJLA and 3FWBC with confidence: 0.981427868693 | SUS1 URM1 |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 2A4VA and 3FWBC with confidence: 0.902997204656 | SUS1 gi|93278640 |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 2C5KT and 3FWBC with confidence: 0.90532765371 | SUS1 gi|88192563, gi|... |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 1QSDA and 3FWBC with confidence: 0.907001717537 | RBL2 SUS1 |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 3FWBC and 3B5NC with confidence: 0.914556977194 | SUS1 gi|160877908, gi... |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 3FWBC and 1USUB with confidence: 0.93372930088 | SUS1 gi|42543757 |