













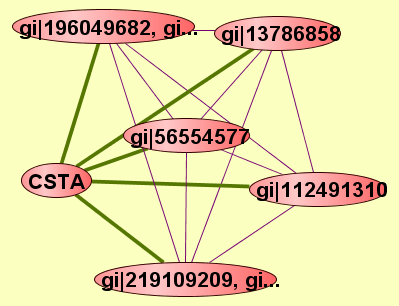
| LEGEND: |
|||
| = Same process, or one is unknown. | = Same branch, distance 4. | ||
| = Same branch, distance 1. | = Same branch, distance 5. | ||
| = Same branch, distance 2. | = Same branch, distance 6 or more. | ||
| = Same branch, distance 3. | = Not in same branch of GO. | ||
| Size | Notes | Members | |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 2VYIA and 1NB5I with confidence: 0.982633832714 | CSTA gi|196049681, gi... |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 2HE4A and 1NB5I with confidence: 0.986319551731 | CSTA gi|112491310 |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 1I1RB and 1NB5I with confidence: 0.920109410039 | CSTA gi|13786858 |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 1XJDA and 1NB5I with confidence: 0.915377225342 | CSTA gi|56554577 |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 2ZHNA and 1NB5I with confidence: 0.911186004394 | CSTA gi|219109210, gi... |