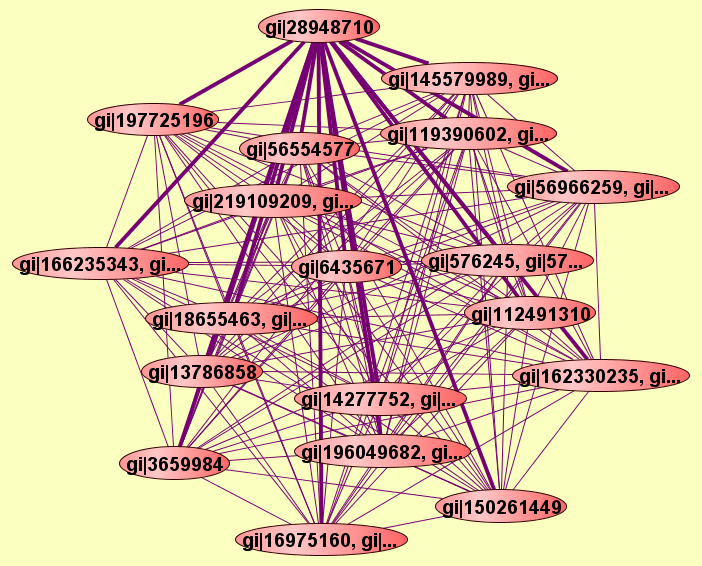
| |
Size |
Notes |
Members |
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1HD2A and 1N1JA with confidence: 0.914988939122 |
gi|16975162, gi|...
gi|28948710
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1BD2D and 1N1JA with confidence: 0.970807364863 |
gi|28948710
gi|3659984
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2VYIA and 1N1JA with confidence: 0.924497023523 |
gi|196049681, gi...
gi|28948710
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2FFQA and 1N1JA with confidence: 0.930549726472 |
gi|110591381, gi...
gi|28948710
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2HE4A and 1N1JA with confidence: 0.998342513278 |
gi|112491310
gi|28948710
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2Q3HA and 1N1JA with confidence: 0.99089187817 |
gi|150261449
gi|28948710
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1N1JA and 1I1RB with confidence: 0.987665976361 |
gi|13786858
gi|28948710
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1N1JA and 1BYGA with confidence: 0.953294714629 |
gi|28948710
gi|6435671
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1N1JA and 3D85A with confidence: 0.955558305226 |
gi|197725196
gi|28948710
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1N1JA and 3B8ZA with confidence: 0.956755150897 |
gi|162330234, gi...
gi|28948710
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1N1JA and 2NS2A with confidence: 0.919875771684 |
gi|119390601, gi...
gi|28948710
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1N1JA and 1KRNA with confidence: 0.937451096217 |
gi|28948710
gi|576245, gi|57...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1N1JA and 2OCTA with confidence: 0.909859131999 |
gi|145579988, gi...
gi|28948710
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1N1JA and 1XJDA with confidence: 0.978910782068 |
gi|28948710
gi|56554577
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1N1JA and 1K1FA with confidence: 0.910027655014 |
gi|18655462, gi|...
gi|28948710
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1N1JA and 1I1JA with confidence: 0.968430887505 |
gi|14277752, gi|...
gi|28948710
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1N1JA and 1UNLA with confidence: 0.974077260348 |
gi|28948710
gi|56966260, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1N1JA and 2ZHNA with confidence: 0.989368242752 |
gi|219109210, gi...
gi|28948710
|