













| General Information: |
|
| Name(s) found: |
CG6262-PB /
FBpp0086295
[FlyBase]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 867 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
trehalose metabolic process
[IEA]
|
| Molecular Function: |
trehalase activity
[ISS]
alpha,alpha-trehalase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHQTARGMID NYLELVQQYG FVPGCGRIYC SGRSNPPLLI MMVKAYVEVT KDEQYAIEAL 60
61 PLLETEYDTF ISKHSVQVKG RTMYQYRDSS AGPRPEAYRE DLECVASIKS PVVREVMYTE 120
121 LKSAAESGTN FSSRWYVTVD GSNKGSFRDI KTSAIVPVEL NCIVFRSGKI LAEFNRKAGN 180
181 TKKADEYQDR ACVLVKAIRD NLWNAQAGIW LDYDLVNNKP RNYFCCTNFA PLWARAFPLV 240
241 DTEKVSKGVM QYIKTNDLDA QYGGVPYTMN KESGQNWDHP NVFPPMMFLI IEGLENLGTP 300
301 PAKAMSKRWA HRWVKSNYAA YKYESFMFEK YYCEDFGTSG GSSPENTPLG YGWTNGVIIE 360
361 FLCKYGKEIS LADTSDEGSK SKSRGSKPSG KYQYIITSEA NMDDQPSKKR CVCGAPSMVG 420
421 TMSHKSTATT ATSDFAQQTQ PPCSCGIAQK QRSQTQPGED GKCSCGAGAK EKSQQQLQPK 480
481 DAPSCGICGS QKQQPSQAQM QRSQQKVDDD QCTCSRKEAL QQSQSQQQRT LGIQHDPDCP 540
541 CAGPQQKGAG LIVVQGQVPD TDPDCECSMD ERQKQQMEQQ QKQQDCMRQQ QQQQQEHQKQ 600
601 QQQLQSEQQQ DCTTKPQTQD CSQHLKKSNG GCGCQDDEDE DRHDALTGHY FPVADKDPDD 660
661 VYQQESKSTQ RESGCDGCGR DDCPDPPQED CGCSPVAKPQ QTPIAECAPQ PKAPQPAPAD 720
721 DCSCAVGKSQ SKAASVVPPP TFICPHCGGV TSEAFLSTHT IGTNRTMGPQ DDCGAAGQPL 780
781 RGLASKGIQS GHSKVVAGGA GAECAPCKAH VTADFPAGHG PKPLFNKQFP LADPASNFDA 840
841 CPPKRGSDPT NNKKKKCCNC DEKEDGE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
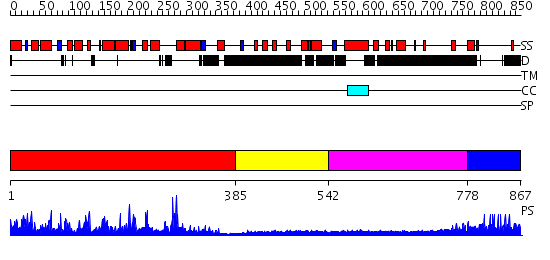
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..384] | 105.0 | No description for 2jf4A was found. | |
| 2 | View Details | [385..541] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [542..777] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [778..867] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)