













| General Information: |
|
| Name(s) found: |
Nup133-PA /
FBpp0083695
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1200 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: |
protein binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MERNLQKQLY GISRESSPGA RRYSMPAASA DSTRKSIFGG SASCAQMSGS LKRTALTNSR 60
61 LSLSVRSTQS IAGIRSDYNS VESFGCPLPV VVNEALTFAG PGAGTVTAKV TQNGWAWVVQ 120
121 GRRLLIWQYK DTAKSGSPPR VGKLARRGGG LAQCRELTLP YSDLGHKSDL ISVFQTEGQQ 180
181 MASCIAVSAT GEVRYWSSIA HDGNSVDLSI LTGQEFVQLL SLPTQQGYLA VTTTCNLVFL 240
241 RVGLTNGRYT LHHKTIKPAT SFLGGFGKKF ASILIGMNTG ADKDQTLVGM CCESNLESGE 300
301 TIVAVLSDRA IQRWSLSNNG NTENLLYEDA DMLRRIREEF ITNFWKFRLP ADSLEIDLHL 360
361 LDFHVVKNKA YILAGAVNAA HAPQMCYALV TGTAQAERML LESFTPLNMN KFFSAKTEED 420
421 CLSVRFVVGS SHIYLYTSKV VYPLHLTNSV PTAELEAEKI EFHQHDDRIL SAVICSQLPL 480
481 FFSRTHGLVS ITPGDFDGTE MMNMSSCNTP DLYAPNSCNA SFAVADHSAL TNSTNNLHLF 540
541 ELDPDEMYNE LSDEVGQLKA AFLYHMKRNS NMVKTIVDEL LRNVTAADPS GAPMDAYKLD 600
601 RIVITIAEDL AEDIPIADPR WEEALADQEM NRHAIGSSRS MQIINQLRDK IIAFQHFITF 660
661 LHSSLVWDKL NVIPCGSHSL KPTGCILADI SEKIVAAMAL RSIQTKLPKL IEEAIDATVA 720
721 LWHEEPQGSL TYQDIFYVKL SRFQNVFEAL ADIADDRIAA QNQTTISVAH FVNEINSIVL 780
781 DVLGQVFKYR KQHASSFRLS HEKLPSYENL PWTAMAGSAG VRDTLTRLID ISVRYGSHCV 840
841 SETELKQQLY QQIFELIDLV LDGRKTYLKS VRDTEKFNVL QQQFEAQRRE LISVLIKDRQ 900
901 YEYAAKIAEK YLDFQSLVLI CDETQDKERL EDYTRKYEEY DFSQFAINWH LRQNRHGEVF 960
961 ERFKGNQTAL AQFMRDHPSL GWIQLIFNGD FERAAKVLYE LAQCETEFVA RKKSMLSLAK1020
1021 LAAFAAAESD LTAQVEKINA DLTLVEYQSQ LGHDVLESFG FDPAEQKVLK AEEIISLYIA1080
1081 EENETASETE FRKALELLSY VEQPYDMRHK IWCAAIKRDN WTDYDPNNAV HYMQKLLFYK1140
1141 IIEISQLMGN DSENVLPPME DFLESVELGD LPQQKPFQYL LKLTYEYVAD MFKQPDDMEL1200
1201 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
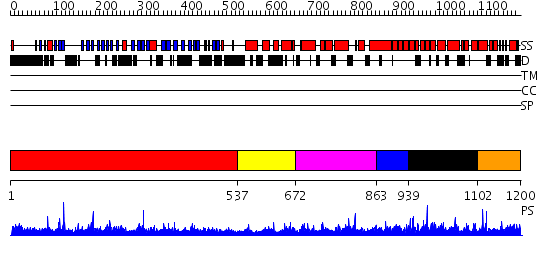
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..536] | 121.0 | The crystal structure of the N-terminal domain of Nup133 reveals a beta-propeller fold common to several nucleoporins | |
| 2 | View Details | [537..671] | 2.068995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [672..862] | 1.07699 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [863..938] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [939..1101] | 2.080982 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [1102..1200] | 1.076985 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
|||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. | |||||||||||||||
| 5 | No functions predicted. | |||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)