













| General Information: |
|
| Name(s) found: |
FBpp0301767
[FlyBase]
FBpp0301766 [FlyBase] tay-PA / FBpp0073960 [FlyBase] |
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2486 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
adult walking behavior
[IMP]
pattern orientation [IMP] |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDTSNASAKS KTSASGSSSI NGIGSQIKHI SSSQDIGSSV PAVPVQDEKS ESHSHSHSHS 60
61 SSSSQSSQSQ SQSQPQSTVL TVVGQSISKS STGALAVLHL PPPTATALVQ ASANIADLGS 120
121 MPRARPSFVS NTSSTAATTT TATSASDGGK TMAVAAIPPA SPVSISEPTC PSVSTVSMAP 180
181 SACDFDSDTE LSIVAAAAAV SASAGIDSRS PTSPPTAAGS TIMPQLNSSA GGTASIAGNQ 240
241 SGFGAGSGAV AAGNTNGHST TSSSSTHLSL NAKQQRQQKR RRERAQRLQA ERDSRCNANS 300
301 LSGTFIAGSS VGVGSAMSAA NGGQGGGVGL VTPNSSVGGG SSAVAMGNGS AAGPGNNGMI 360
361 YHINSAGSMG EDSNNSNGNF TSSSNGSNNL LPPTDSIASL LLNPPHSPEC NSGLMATPSD 420
421 SEGESLDEDL LSTSSSSTLL LPRDKPPPRP PPPVRRKLPR SPVLEEEIID GFAILEFKTY 480
481 EDLEFAIKLG QKRKEKRLSA LDELTCTYSI EEMKIPKVID TGQSQPLRVT SLSTLTVNNN 540
541 NLKENPQPSL HRDRGGNVIS NNNNNNSSNN NNSMINSNGP NANSNSSSLS NVVYLLPSVN 600
601 MVGNVLETEA EPRDHWRSEY GQQQDQLQSD NSANSKSNNN ISDDNQINNS SHQINHTTNN 660
661 QSSKQQQAAD GGCNAAKMQV NNSATVSTRF LENDNDSLML DISLPSLGVG GAVSHSHDHI 720
721 DVGVIHASCT GSSSDTKKSH NSPATSNTID QLHVTKPLPT HQLNNLISSD LNAAANANAN 780
781 SNHLGVCKLG SQSAQVAKKS MGLENIETVT NSATCRSLDI QELANVSAQS FGASSSSSSS 840
841 SISIKKENEQ SDLLNDASTN CSSNSKGNNI CDNVSNDKKN SEQIFDIDNP PTPMNISNTS 900
901 SCSDRSSSRK FSELPHLKKK SAACGDRDNG PYAGVPFAAA MGTVSPDSDP SKALNIKNAS 960
961 ESASGKAVHA HAGAPIPPPP AASVGDPGNR ELGVLQKTSP DKPLYPNLCA PPSVSSVQFP1020
1021 NVGISAGNEK IASTVASDID QKIIFNSKAT ASVSNGLSSG VQRSIVPLLS AAPSPAAVTL1080
1081 SSPFSAHIAA APNAAGLSIL SSASSVILAN SSIKNTNNGS NNGSANGQGQ LSSVGLSSTS1140
1141 EIAKSNSVPT TSSPPNPMVN GFLSVYPPPA ATIATSSVTT SALSRVSPNA ANKLVPPVSA1200
1201 TTSISINTNY VGISGGTTTT SGMGKVVFGA GGNAIVGTGS PLLFPGAPPA PISHVATGVP1260
1261 MIIQAPPYRA PYTNYPLYTP YSGLTHGSYL PPVLPVATPN LSNLPPTQHR SSDSRNSRES1320
1321 PASLKSTPSN IGLNVSMAPT LRSITPLNNS SAISSGASQP VVSVVPSANS TALSMSNPHI1380
1381 SHSHHVPAYA SGAFSSSAAA GTSTPNSGLS TLAVTSLSTS AAPQPHSHFP QSTQMLPQSG1440
1441 NFSSVSHLLT THPMSSQNQP MVRCGSTLYS QSSAAATAPP SAAAAVSNFT PSVLAVQSLT1500
1501 TAVTSSSSSP STLSSSVIQK VISPKGESPC NKDRDSSYST NIRSHGASTM LPGASQGIGL1560
1561 GSSFCGIAPS QYGGTGTPVL SSSSSMPPGL GNLSSYSSKA ALWLNSPANA VVTTCAPTTP1620
1621 IVSSGSARPT PPLSNCTSMG IGMVNAASTA RSSCNAISPL SIPATAGIHV SATNPSFQSS1680
1681 SYFPTPLAPP PSSPSPATSS AAIISSSASQ FNPAVSHSMS SIVTTAGATT TTASSVTQPS1740
1741 VAAISNPVTN TPHPFSAESL FQPSKNDQAD LLRRELDSRF LDRSGLAVTP TPPSSTYLRT1800
1801 DLQHQQHAHL HQHPQLLPPV SAPSTPLSAV QPSSGQIFPP PLFKDISKIS SVDPQFYRTG1860
1861 MGLPPGGYSG YTSAGLLHSG LGGPTPFMPP NHLTSFAPKK TGRWNAMHVR IAWEIYYHQN1920
1921 KQSPEKQPGF PTASSNAGSI NAPIPPGNMI SGGGGGMASG ASTPVNNVAA IPAVGGGVLV1980
1981 SNGPPSVLGM KTTPTLGMSS TPQHILHRAS EMPPSAAFPG GLPGRLAFET SPLAASFIGA2040
2041 PPSHIGTAVS PFGRYVTPFG FTGLTHFGGH LDSWRTNAMQ RSVAYHPSSA APSPNWPVKS2100
2101 DPGLDNARRE AEERERELRD REQRERDRQR REREERERKE KEEKLKREQQ ERERERERDR2160
2161 KERERERREI ERREMERERM LQQQRINESN KQAAVQAAAA ATAPVRDRSP HRNVGELNTE2220
2221 IRIKEEHPRT KEEQDVMLLR ASAAAVGVGD PRYHSSSLAA AQASAAAAAA HHHHANFMAA2280
2281 SRHGLPPPPS HLTRTMMPPT LSVGGPITHF GAPAPPGWGI DPYRDPYPIL RYNPIMEAAL2340
2341 RHEAEERQKA INIYAAQSAA HLRSKEPSPI PPSVGSLGPP PPHHRLQIVS SVAQSGVQQP2400
2401 GPPSQQQQQQ MSSGGGMVKN SGGPQAPGSI PVQHMISVDS MGQQSVSAKK ESDHTMGIVS2460
2461 NAGVGLSSVP GGVIGISPVS SVAPSR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
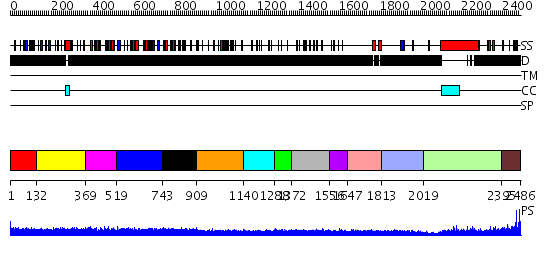
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..131] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [132..368] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [369..518] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [519..742] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [743..908] | 1.101981 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [909..1139] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1140..1287] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1288..1371] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1372..1555] | 2.090985 | View MSA. No confident structure predictions are available. | |
| 10 | View Details | [1556..1646] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1647..1812] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1813..2018] | N/A | No confident structure predictions are available. | |
| 13 | View Details | [2019..2394] | 30.39794 | Heavy meromyosin subfragment | |
| 14 | View Details | [2395..2486] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.68 |
Source: Reynolds et al. (2008)