













| General Information: |
|
| Name(s) found: |
ASPP-PA /
FBpp0071548
[FlyBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1020 amino acids |
Gene Ontology: |
|
| Cellular Component: |
adherens junction
[IDA]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
protein tyrosine kinase activator activity
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKEPTNTLDE IVPSRLTSAE LRAMALRQQQ QIDSQHQLLA TKEQRLRFLK SQEVRSAVAS 60
61 AEGERLRRLR ERVEAQESKL RRLRALRGQV DLQKTYNVTL SNDLDSIRAL FSEKEKELSL 120
121 AVAKVEALTR QLEELRRDRR CPVNILTAAG NGVGGQSLPP QASRELEKLR RELMYRNQLS 180
181 LQQDARLHMQ REALQQRQAE LRSTDQRIYE LQTRLQRKKQ ANTHHQQQQQ QQQQHQQQQH 240
241 QQQHQQTTVQ QQQQQQQQLH VQSAQLLAAA AAATAQQQQQ HQQHQQQQQQ QQQQSASNLG 300
301 KHGGVAALKQ QLLQKQVNQL AAAAAAAAAG RARGNVAAVE PFIHTPQKST ITSTASYLSG 360
361 GMKHAAATAN TNQQNQLIQD LTHVKLGQNS GFFAGSVPVP VASSSCESDE SKLAKKLLAA 420
421 PAKTEAELRK QAQTEAMDST THIYAEVGPK KRDREAAAAA AAAAAAAAAA AQAAAEAANQ 480
481 AATAAAASAI SPPATANASK IPKSTTSSSS SASALISKLN QQATAKESQD QQQSQAKKNE 540
541 ISVTSETLSE RNTQQQLTVH SMPLGAISKS VALAVSNASK PLQVQVSNVA VPPRKPISSV 600
601 APNSVSSGSS IPKMITYSPK VNRVAPNVVM TDPRPALPPK PSKMSPTEQP SPVEANTSGS 660
661 AGGAGSGAAS ASTANKTQTA TFGLQSLNIN DNLPIKAKPL TIRKQPLFEQ PRLKSSQSSS 720
721 NGQQKPPLLQ LQNQRKSEPS ARQPTENLPD TSPQHSPSSE SSADETDRMV APSITTSQQE 780
781 TSTSATSTTT TVTTSNIKER TGNGKPKLGR RVSFDPLALL LDASLEGELE LVKKTAMQVA 840
841 NPSAANDEGI TALHNAICAG HIDIVKFLVE FGCDVNAQDS DGWTPLHCAA SCNNLSMVKF 900
901 LVESGACLFA STLSDHETPA EKCEEDEEGF DGCSEYLYSI QEKLGILHNG DVYAVFSYEA 960
961 QNGDELSFHV NEPLIVLRKG DDAENEWWWA RNAAGEEGYV PRNLLGLYPR VPPHSPHFSD1020
1021 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
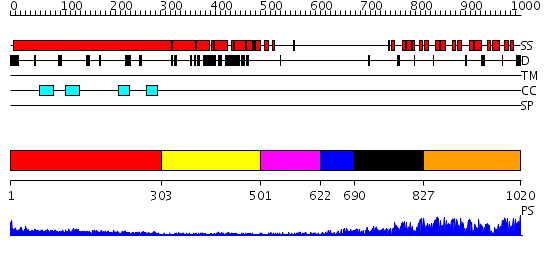
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..302] | 21.0 | Heavy meromyosin subfragment | |
| 2 | View Details | [303..500] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [501..621] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [622..689] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [690..826] | 12.221849 | Crystal structure of UPLC1 GAP domain | |
| 6 | View Details | [827..1020] | 40.522879 | 53BP2 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)