













| General Information: |
|
| Name(s) found: |
FBpp0304114
[FlyBase]
FBpp0304113 [FlyBase] FBpp0304112 [FlyBase] Trf4-1-PE / FBpp0071159 [FlyBase] Trf4-1-PC / FBpp0071158 [FlyBase] |
| Description(s) found:
Found 47 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1001 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
sister chromatid cohesion
[NAS]
ncRNA polyadenylation involved in polyadenylation-dependent ncRNA catabolic process [IMP] |
| Molecular Function: |
DNA-directed DNA polymerase activity
[NAS]
DNA topoisomerase activity [ISS] polynucleotide adenylyltransferase activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDPFVGWFQK EQEGPSLCTW LKIWETNAQE MGALLNQQLQ QATTINGSTS SSSSSSNSGN 60
61 NNNNNNNNII NNTITNTTNN TGNNSSAKPY LSRPYSSLNR VLNFRADSLE ILQQQQQQQQ 120
121 LNGTTQRNST NINTTSGGST SSSADSTTNR DNNSPANSSS TNGPGAGTGT STGAGGTGTN 180
181 SPATTASSTA ATTTGPATSM SDTSNNPPQS TTTPASRTNS IYYNPSRKKR PENKAGGAHY 240
241 YMNNHMEMIA KYKGEPWRKP DYPYGEGVIG LHEEIEHFYQ YVLPTPCEHA IRNEVVKRIE 300
301 AVVHSIWPQA VVEIFGSFRT GLFLPTSDID LVVLGLWEKL PLRTLEFELV SRGIAEACTV 360
361 RVLDKASVPI IKLTDRETQV KVDISFNMQS GVQSAELIKK FKRDYPVLEK LVLVLKQFLL 420
421 LRDLNEVFTG GISSYSLILM CISFLQMHPR GIYHDTANLG VLLLEFFELY GRRFNYMKIG 480
481 ISIKNGGRYM PKDELQRDMV DGHRPSLLCI EDPLTPGNDI GRSSYGVFQV QQAFKCAYRV 540
541 LALAVSPLNL LGIDPRVNSI LGRIIHITDD VIDYREWIRE NFEHLVVVDR ISPLPTAAPT 600
601 AYATANGAPK YVIMPSGAVV QQLYHHPVQV PTAHGHSHAH SHSHGHAHPG AHLCQPYVTG 660
661 TTVSAVTTTT TMAVVTVGVS AGGVQQQQQQ QNATAHTHSQ QQTQNQSQSR HRRGSTSSGD 720
721 DSEDSKDGDV VETTSSAQEV VDIALSTPNG LANMSMPMPV HAVGMPASNS WSGNGNGNGN 780
781 SSSSTGSSPE IAHIAAQEMD PELEDQQQQQ QHQETSGGNG FIRPGDVGTG SNRGGGDGSG 840
841 GRNYNQRNNH NSSGYYHQQY YVPPPMQQQL SKSNSSSNYH QQHHHSHSHG NHSHRQQHHH 900
901 QQQHHHQQRP QHLRVGGGNR YQKSLGGSPI ISAGNASNSS SNCSNSSSSS GSNNSRLPPL 960
961 RGTLVNSSSA ISIISISSES SIASSSSSSS RSGQDQQRDE R |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
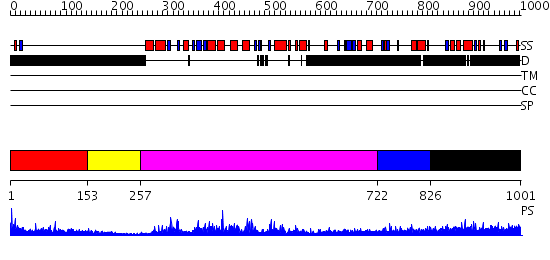
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..152] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [153..256] | 29.522879 | Structural Basis for UTP Specificity of RNA Editing TUTases From Trypanosoma Brucei | |
| 3 | View Details | [257..721] | 93.69897 | Crystal structure of poly(A) polymerase in complex with 3'-dATP and magnesium chloride | |
| 4 | View Details | [722..825] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [826..1001] | 5.065995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 |
|
|||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.81 |
Source: Reynolds et al. (2008)