













| General Information: |
|
| Name(s) found: |
AGAP7
[HGNC (HUGO)]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 663 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGNILTCRVH PSVSLEFDQQ QGSVCPSESE IYEAGAEDRM AGAPMAAAVQ PAEVTVEVGE 60
61 DLHMHHVRDR EMPEALEFNP SANPEASTIF QRNSQTDVVE IRRSNCTNHV STVHFSQQYS 120
121 LCSTIFLDDS TAIQHYLTMT IISVTLEIPH HITQRDADRS LSIPDEQLHS FAVSTVHITK 180
181 NRNGGGSLNN YSSSIPSTPS TSQEDPQFSV PPTANTPTPV CKRSMRWSNL FTSEKGSDPD 240
241 KERKAPENHA DTIGSGRAIP IKQGMLLKRS GKWLKTWKKK YVTLCSNGVL TYYSSLGDYM 300
301 KYIHKKEIDL QTSTIKVPGK WPSLATLACT PISSSKSDGL SKDMDTGLGD SICFSPSISS 360
361 TTIPKLNPPP SPHANKKKHL KKKSTNNFMI VSATGQTWHF EATTYEERDA WVQAIQSQIL 420
421 ASLQSCESSK SKSQLTSQSE AMALQSIQNM RGNAHCVDCE TQNPKWASLN LGVLMCIECS 480
481 GIHRSLGTRL SRVRSLELDD WPVELRKVMS SIGNDLANSI WEGSSQGRTK PTEKSTREEK 540
541 ERWIRSKYEE KLFLAPLPCT ELSLGQQLLR ATADEDLQTA ILLLAHGSRE EVNETCGEGD 600
601 GCTALHLACR KGNVVLAQLL IWYGVDVMAR DAHGNTALTY ARQASSQECI NVLLQYGCPD 660
661 ECV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
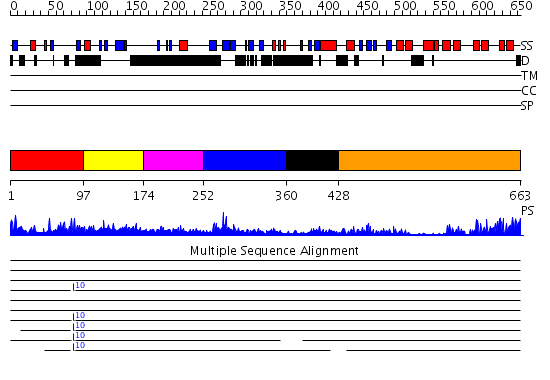
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..96] | 2.028992 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [97..173] | 7.0 | Centaurin Gamma 1 (Human) | |
| 3 | View Details | [174..251] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [252..359] | 15.39794 | Triglycine variant of the ARNO Pleckstrin Homology Domain in complex with Ins(1,3,4,5)P4 | |
| 5 | View Details | [360..427] | 1.52 | Crystal Structure of the PH Domain of SKAP-Hom with 8 Vector-derived N-terminal Residues | |
| 6 | View Details | [428..663] | 27.0 | Pyk2-associated protein beta; Pyk2-associated protein beta ARF-GAP domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)