













| General Information: |
|
| Name(s) found: |
ASPSCR1
[HGNC (HUGO)]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Homo sapiens |
| Length: | 553 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAPAGGGGS AVSVLAPNGR RHTVKVTPST VLLQVLEDTC RRQDFNPCEY DLKFQRSVLD 60
61 LSLQWRFANL PNNAKLEMVP ASRSREGPEN MVRIALQLDD GSRLQDSFCS GQTLWELLSH 120
121 FPQIRECLQH PGGATPVCVY TRDEVTGEAA LRGTTLQSLG LTGGSATIRF VMKCYDPVGK 180
181 TPGSLGSSAS AGQAAASAPL PLESGELSRG DLSRPEDADT SGPCCEHTQE KQSTRAPAAA 240
241 PFVPFSGGGQ RLGGPPGPTR PLTSSSAKLP KSLSSPGGPS KPKKSKSGQD PQQEQEQERE 300
301 RDPQQEQERE RPVDREPVDR EPVVCHPDLE ERLQAWPAEL PDEFFELTVD DVRRRLAQLK 360
361 SERKRLEEAP LVTKAFREAQ IKEKLERYPK VALRVLFPDR YVLQGFFRPS ETVGDLRDFV 420
421 RSHLGNPELS FYLFITPPKT VLDDHTQTLF QANLFPAALV HLGAEEPAGV YLEPGLLEHA 480
481 ISPSAADVLV ARYMSRAAGS PSPLPAPDPA PKSEPAAEEG ALVPPEPIPG TAQPVKRSLG 540
541 KVPKWLKLPA SKR |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
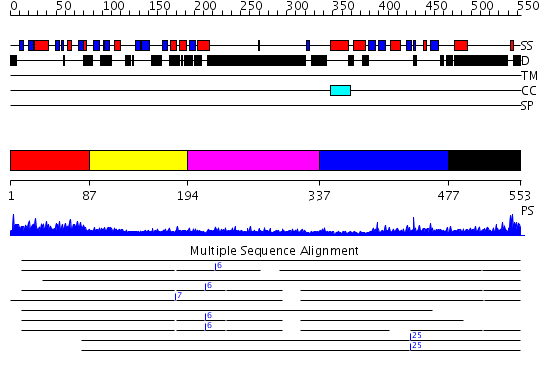
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..86] | 31.0 | No description for 2al3A was found. | |
| 2 | View Details | [87..193] | 0.959 | The deposition and citation title is: Solution structure of the UBX domain of D0H8S2298E protein | |
| 3 | View Details | [194..336] | 3.240995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [337..476] | 15.30103 | Crystal structure of AAA ATPase p97/VCP ND1 in complex with p47 C | |
| 5 | View Details | [477..553] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. | ||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)