













| General Information: |
|
| Name(s) found: |
gi|38229313
[NCBI NR]
gi|167009139 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Homo sapiens |
| Length: | 739 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGAVPCRRAL LLCNGMRYKL LQEGDIQVCV IRHPRTFLSK ILTSKFLRRW EPHHLTLADN 60
61 SLASATPTGY MENSVSYSAI EDVQLLSWEN APKYCLQLTI PGGTVLLQAA NSYLRDQWFH 120
121 SLQWKKKIYK YKKVLSNPSR WEVVLKEIRT LVDMALTSPL QDDSINQAPL EIVSKLLSEN 180
181 TNLTTQEHEN IIVAIAPLLE NNHPPPDLCE FFCKHCRERP RSMVVIEVFT PVVQRILKHN 240
241 MDFGKCPRLR LFTQEYILAL NELNAGMEVV KKFIQSMHGP TGHCPHPRVL PNLVAVCLAA 300
301 IYSCYEEFIN SRDNSPSLKE IRNGCQQPCD RKPTLPLRLL HPSPDLVSQE ATLSEARLKS 360
361 VVVASSEIHV EVERTSTAKP ALTASAGNDS EPNLIDCLMV SPACSTMSIE LGPQADRTLG 420
421 CYVEILKLLS DYDDWRPSLA SLLQPIPFPK EALAHEKFTK ELKYVIQRFA EDPRQEVHSC 480
481 LLSVRAGKDG WFQLYSPGGV ACDDDGELFA SMVHILMGSC YKTKKFLLSL AENKLGPCML 540
541 LALRGNQTMV EILCLMLEYN IIDNNDTQLQ IISTLESTDV GKRMYEQLCD RQRELKELQR 600
601 KGGPTRLTLP SKSTDADLAR LLSSGSFGNL ENLSLAFTNV TSACAEHLIK LPSLKQLNLW 660
661 STQFGDAGLR LLSEHLTMLQ VLNLCETPVT DAGLLALSSM KSLCSLNMNS TKLSADTYED 720
721 LKAKLPNLKE VDVRYTEAW |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
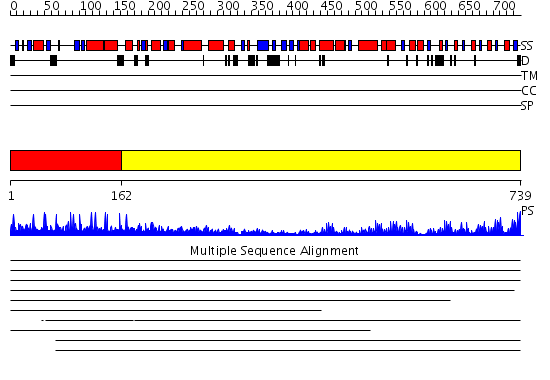
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..161] | 1.0 | Solution structure of the PH domain of human Docking protein BRDG1 | |
| 2 | View Details | [162..739] | 24.0 | The molecular structure of toll-like receptor 3 ligand binding domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)