













| General Information: |
|
| Name(s) found: |
gi|38201698
[NCBI NR]
gi|22759954 [NCBI NR] gi|119628575 [NCBI NR] |
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 807 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPRAPRCRAV RSLLRSHYRE VLPLATFVRR LGPQGWRLVQ RGDPAAFRAL VAQCLVCVPW 60
61 DARPPPAAPS FRQVSCLKEL VARVLQRLCE RGAKNVLAFG FALLDGARGG PPEAFTTSVR 120
121 SYLPNTVTDA LRGSGAWGLL LRRVGDDVLV HLLARCALFV LVAPSCAYQV CGPPLYQLGA 180
181 ATQARPPPHA SGPRRRLGCE RAWNHSVREA GVPLGLPAPG ARRRGGSASR SLPLPKRPRR 240
241 GAAPEPERTP VGQGSWAHPG RTRGPSDRGF CVVSPARPAE EATSLEGALS GTRHSHPSVG 300
301 RQHHAGPPST SRPPRPWDTP CPPVYAETKH FLYSSGDKEQ LRPSFLLSSL RPSLTGARRL 360
361 VETIFLGSRP WMPGTPRRLP RLPQRYWQMR PLFLELLGNH AQCPYGVLLK THCPLRAAVT 420
421 PAAGVCAREK PQGSVAAPEE EDTDPRRLVQ LLRQHSSPWQ VYGFVRACLR RLVPPGLWGS 480
481 RHNERRFLRN TKKFISLGKH AKLSLQELTW KMSVRDCAWL RRSPGVGCVP AAEHRLREEI 540
541 LAKFLHWLMS VYVVELLRSF FYVTETTFQK NRLFFYRKSV WSKLQSIGIR QHLKRVQLRE 600
601 LSEAEVRQHR EARPALLTSR LRFIPKPDGL RPIVNMDYVV GARTFRREKR AERLTSRVKA 660
661 LFSVLNYERA RRPGLLGASV LGLDDIHRAW RTFVLRVRAQ DPPPELYFVK VDVTGAYDTI 720
721 PQDRLTEVIA SIIKPQNTYC VRRYAVVQKA AHGHVRKAFK SHVLRPVPGD PAGLHPLHAA 780
781 LQPVLRRHGE QAVCGDSAGR AAPAFGG |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
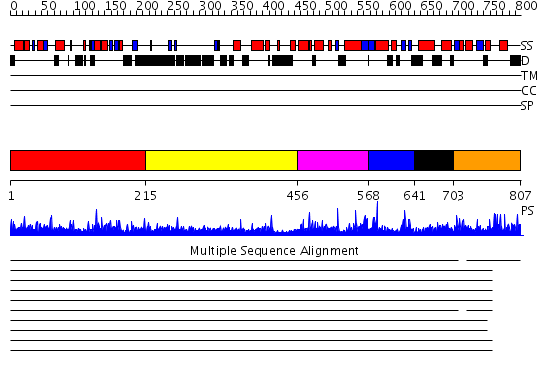
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..214] | 1.02 | Crystal Structure of the TEN domain of the Telomerase Reverse Transcriptase | |
| 2 | View Details | [215..455] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [456..567] | 6.083993 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [568..640] | 3.083991 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [641..702] | 3.079985 | View MSA. Confident ab initio structure predictions are available. | |
| 6 | View Details | [703..807] | 6.075994 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)