













| General Information: |
|
| Name(s) found: |
COPB1
[HGNC (HUGO)]
|
| Description(s) found:
Found 41 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 953 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTAAENVCYT LINVPMDSEP PSEISLKNDL EKGDVKSKTE ALKKVIIMIL NGEKLPGLLM 60
61 TIIRFVLPLQ DHTIKKLLLV FWEIVPKTTP DGRLLHEMIL VCDAYRKDLQ HPNEFIRGST 120
121 LRFLCKLKEA ELLEPLMPAI RACLEHRHSY VRRNAVLAIY TIYRNFEHLI PDAPELIHDF 180
181 LVNEKDASCK RNAFMMLIHA DQDRALDYLS TCIDQVQTFG DILQLVIVEL IYKVCHANPS 240
241 ERARFIRCIY NLLQSSSPAV KYEAAGTLVT LSSAPTAIKA AAQCYIDLII KESDNNVKLI 300
301 VLDRLIELKE HPAHERVLQD LVMDILRVLS TPDLEVRKKT LQLALDLVSS RNVEELVIVL 360
361 KKEVIKTNNV SEHEDTDKYR QLLVRTLHSC SVRFPDMAAN VIPVLMEFLS DNNEAAAADV 420
421 LEFVREAIQR FDNLRMLIVE KMLEVFHAIK SVKIYRGALW ILGEYCSTKE DIQSVMTEIR 480
481 RSLGEIPIVE SEIKKEAGEL KPEEEITVGP VQKLVTEMGT YATQSALSSS RPTKKEEDRP 540
541 PLRGFLLDGD FFVAASLATT LTKIALRYVA LVQEKKKQNS FVAEAMLLMA TILHLGKSSL 600
601 PKKPITDDDV DRISLCLKVL SECSPLMNDI FNKECRQSLS HMLSAKLEEE KLSQKKESEK 660
661 RNVTVQPDDP ISFMQLTAKN EMNCKEDQFQ LSLLAAMGNT QRKEAADPLA SKLNKVTQLT 720
721 GFSDPVYAEA YVHVNQYDIV LDVLVVNQTS DTLQNCTLEL ATLGDLKLVE KPSPLTLAPH 780
781 DFANIKANVK VASTENGIIF GNIVYDVSGA ASDRNCVVLS DIHIDIMDYI QPATCTDAEF 840
841 RQMWAEFEWE NKVTVNTNMV DLNDYLQHIL KSTNMKCLTP EKALSGYCGF MAANLYARSI 900
901 FGEDALANVS IEKPIHQGPD AAVTGHIRIR AKSQGMALSL GDKINLSQKK TSI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
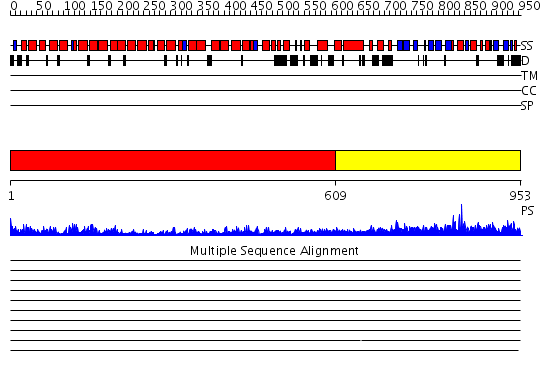
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..608] | 83.39794 | AP1 clathrin adaptor core | |
| 2 | View Details | [609..953] | 69.69897 | Structural Identification of a conserved appendage domain in the carboxyl-terminus of the COPI gamma-subunit. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)