













| General Information: |
|
| Name(s) found: |
gi|60459550
[NCBI NR]
gi|26638662 [NCBI NR] gi|119623925 [NCBI NR] gi|119623915 [NCBI NR] gi|119623912 [NCBI NR] |
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 851 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASLGANPRR TPQGPRPGAA SSGFPSPAPV PGPREAEEEE VEEEEELAEI HLCVLWNSGY 60
61 LGIAYYDTSD STIHFMPDAP DHESLKLLQR VLDEINPQSV VTSAKQDENM TRFLGKLASQ 120
121 EHREPKRPEI IFLPSVDFGL EISKQRLLSG NYSFIPDAMT ATEKILFLSS IIPFDCLLTP 180
181 PGDLRFTPIP LLIPSQVRAL GGLLKFLGRR RIGVELEDYN VSVPILGFKK FMLTHLVNID 240
241 QDTYSVLQIF KSESHPSVYK VASGLKEGLS LFGILNRCHC KWGEKLLRLW FTRPTHDLGE 300
301 LSSRLDVIQF FLLPQNLDMA QMLHRLLGHI KNVPLILKRM KLSHTKVSDW QVLYKTVYSA 360
361 LGLRDACRSL PQSIQLFRDI AQEFSDDLHH IASLIGKVVD FEGSLAENRF TVLPNIDPEI 420
421 DEKKRRLMGL PSFLTEVARK ELENLDSRIP SCSVIYIPLI GFLLSIPRLP SMVEASDFEI 480
481 NGLDFMFLSE EKLHYRSART KELDALLGDL HCEIRDQETL LMYQLQCQVL ARAAVLTRVL 540
541 DLASRLDVLL ALASAARDYG YSRPRYSPQV LGVRIQNGRH PLMELCARTF VPNSTECGGD 600
601 KGRVKVITGP NSSGKSIYLK QVGLITFMAL VGSFVPAEEA EIGAVDAIFT RIHSCESISL 660
661 GLSTFMIDLN QVAKAVNNAT AQSLVLIDEF GKGTNTVDGL ALLAAVLRHW LARGPTCPHI 720
721 FVATNFLSLV QLQLLPQGPL VQYLTMETCE DGNDLVFFYQ VCEGVAKASH ASHTAAQAGL 780
781 PDKLVARGKE VSDLIRSGKP IKPVKDLLKK NQMENCQTLV DKFMKLDLED PNLDLNVFMS 840
841 QEVLPAATSI L |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
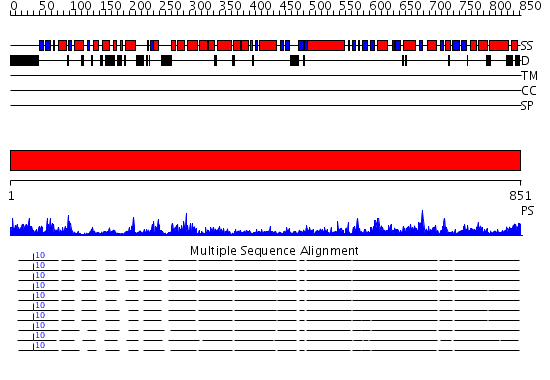
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..851] | 161.0 | Crystal Structure of E. coli DNA Mismatch Repair enzyme MutS, E38T mutant, in complex with a G.T mismatch |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)