













| General Information: |
|
| Name(s) found: |
NUP133
[HGNC (HUGO)]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Homo sapiens |
| Length: | 1156 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFPAAPSPRT PGTGSRRGPL AGLGPGSTPR TASRKGLPLG SAVSSPVLFS PVGRRSSLSS 60
61 RGTPTRMFPH HSITESVNYD VKTFGSSLPV KVMEALTLAE VDDQLTINID EGGWACLVCK 120
121 EKLIIWKIAL SPITKLSVCK ELQLPPSDFH WSADLVALSY SSPSGEAHST QAVAVMVATR 180
181 EGSIRYWPSL AGEDTYTEAF VDSGGDKTYS FLTAVQGGSF ILSSSGSQLI RLIPESSGKI 240
241 HQHILPQGQG MLSGIGRKVS SLFGILSPSS DLTLSSVLWD RERSSFYSLT SSNISKWELD 300
301 DSSEKHAYSW DINRALKENI TDAIWGSESN YEAIKEGVNI RYLDLKQNCD GLVILAAAWH 360
361 SADNPCLIYY SLITIEDNGC QMSDAVTVEV TQYNPPFQSE DLILCQLTVP NFSNQTAYLY 420
421 NESAVYVCST GTGKFSLPQE KIVFNAQGDS VLGAGACGGV PIIFSRNSGL VSITSRENVS 480
481 ILAEDLEGSL ASSVAGPNSE SMIFETTTKN ETIAQEDKIK LLKAAFLQYC RKDLGHAQMV 540
541 VDELFSSHSD LDSDSELDRA VTQISVDLMD DYPASDPRWA ESVPEEAPGF SNTSLIILHQ 600
601 LEDKMKAHSF LMDFIHQVGL FGRLGSFPVR GTPMATRLLL CEHAEKLSAA IVLKNHHSRL 660
661 SDLVNTAILI ALNKREYEIP SNLTPADVFF REVSQVDTIC ECLLEHEEQV LRDAPMDSIE 720
721 WAEVVINVNN ILKDMLQAAS HYRQNRNSLY RREESLEKEP EYVPWTATSG PGGIRTVIIR 780
781 QHEIVLKVAY PQADSNLRNI VTEQLVALID CFLDGYVSQL KSVDKSSNRE RYDNLEMEYL 840
841 QKRSDLLSPL LSLGQYLWAA SLAEKYCDFD ILVQMCEQTD NQSRLQRYMT QFADQNFSDF 900
901 LFRWYLEKGK RGKLLSQPIS QHGQLANFLQ AHEHLSWLHE INSQELEKAH ATLLGLANME 960
961 TRYFAKKKTL LGLSKLAALA SDFSEDMLQE KIEEMAEQER FLLHQETLPE QLLAEKQLNL1020
1021 SAMPVLTAPQ LIGLYICEEN RRANEYDFKK ALDLLEYIDE EEDININDLK LEILCKALQR1080
1081 DNWSSSDGKD DPIEVSKDSI FVKILQKLLK DGIQLSEYLP EVKDLLQADQ LGSLKSNPYF1140
1141 EFVLKANYEY YVQGQI |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample pIC145 from april 2005 | Okada, M., et al. (2006) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
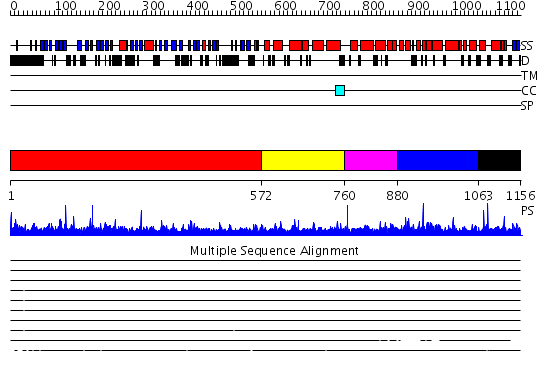
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..571] | 165.0 | The crystal structure of the N-terminal domain of Nup133 reveals a beta-propeller fold common to several nucleoporins | |
| 2 | View Details | [572..759] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [760..879] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [880..1062] | 2.073995 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [1063..1156] | 1.068984 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
|||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. | |||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.72 |
Source: Reynolds et al. (2008)