













| General Information: |
|
| Name(s) found: |
SYCP2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 856 amino acids |
Gene Ontology: |
|
| Cellular Component: |
synaptonemal complex
[IDA]
|
| Biological Process: |
reciprocal meiotic recombination
[IMP]
synapsis [IMP] |
| Molecular Function: |
protein binding
[IPI]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQKLGFPAMK SFDQLRSLPG SAKTYFFSTR PPQDSVSSGS FSNLKLTAEK LVKDQAAMRT 60
61 DLELANCKLK KSMEHVYALE EKLQSAFNEN AKLRVRQKED EKLWRGLESK FSSTKTLCDQ 120
121 LTETLQHLAS QVQDAEKDKG FFETKFNTSS EAINSLNQQM RDMSLRLDAA KEEITSRDKE 180
181 LEELKLEKQH KEMFYQTERC GTASLIEKKD AVITELETTA AERKLKIEKL NSQLEKLHLE 240
241 LTTKEDEVIH LVSIQEKLEK EKTNVQLSSD ELFEKLVRSE QEVKKLDELV HYLIAELTEL 300
301 DKKNLTFKEK FDKLSGLYDT HFMLLRKDRD LASDRAQRSF DQLQGELFRV AAEKEALESS 360
361 GNELSEKIVE LQNDKESLIS QLSGVRCSAS QTIDKLEFEA KGLVLKNAET ESVISKLKEE 420
421 IDTLLESVRT SEDKKKELSI KLSSLEIESK DKYEKLQADA QRQVGELETL QKESESHQLQ 480
481 ADLLAKEVNQ LQTIIEEKGH LILQCNENEK NINQQIIKDK ELLATAETKL AEAKKQYDLM 540
541 LESKQLELSR HLKELSQRND QAINEIRRKY DVEKHEIINS EKDKVEKIIK ELSTKYDKGL 600
601 SDCKEESKRQ LLTIQEEHSS RILNIREEHE SKELNLKAKY DQELRQNQIQ AENELKERIT 660
661 ALKSEHDAQL KAFKCQYEDD CKKLQEELDL QRKKEERQRA LVQLQWKVMS DNPPEEQEVN 720
721 SNKDYSHSSV KVKESRLGGN KRSEHITESP FVKAKVTSVS NILKEATNPK HHSKVTHREY 780
781 EVETNNGRIP KRRKTRQTTM FQEPQRRSTR LTPKLMTPTI IAKETAMADH PHSANIGDLF 840
841 SEGSLNPYAD DPYAFD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
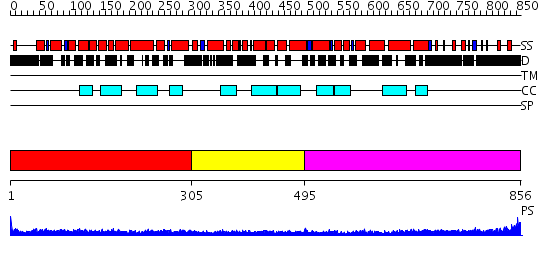
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..304] | 9.69897 | Tropomyosin | |
| 2 | View Details | [305..494] | 2.27 | Ribonuclease domain of colicin E3; Colicin E3 translocation domain; Colicin E3 receptor domain | |
| 3 | View Details | [495..856] | 17.09691 | Heavy meromyosin subfragment |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)