













| General Information: |
|
| Name(s) found: |
XDHA_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 752 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRVDAIAKVT GRARYTDDYV MAGMCYAKYV RSPIAHGYAV SINDEQARSL PGVLAIFTWE 60
61 DVPDIPFATA GHAWTLDENK RDTADRALLT RHVRHHGDAV AIVVARDELT AEKAAQLVSI 120
121 EWQELPVITT PEAALAEDAA PIHNGGNLLK QSTMSTGNVQ QTIDAADYQV QGHYQTPVIQ 180
181 HCHMESVTSL AWMEDDSRIT IVSSTQIPHI VRRVVGQALD IPWSCVRVIK PFVGGGFGNK 240
241 QDVLEEPMAA FLTSKLGGIP VKVSLSREEC FLATRTRHAF TIDGQMGVNR DGTLKGYSLD 300
301 VLSNTGAYAS HGHSIASAGG NKVAYLYPRC AYAYSSKTCY TNLPSAGAMR GYGAPQVVFA 360
361 VESMLDDAAT ALGIDPVEIR LRNAAREGDA NPLTGKRIYS AGLPECLEKG RKIFEWEKRR 420
421 AECQNQQGNL RRGVGVACFS YTSNTWPVGV EIAGARLLMN QDGTINVQSG ATEIGQGADT 480
481 VFSQMVAETV GVPVSDVRVI STQDTDVTPF DPGAFASRQS YVAAPALRSA ALLLKEKIIA 540
541 HAAVMLHQSA MNLTLIKGHI VLVERPEEPL MSLKDLAMDA FYHPERGGQL SAESSIKTTT 600
601 NPPAFGCTFV DLTVDIALCK VTINRILNVH DSGHILNPLL AEGQVHGGMG MGIGWALFEE 660
661 MIIDAKSGVV RNPNLLDYKM PTMPDLPQLE SAFVEINEPQ SAYGHKSLGE PPIIPVAAAI 720
721 RNAVKMATGV AINTLPLTPK RLYEEFHLAG LI |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
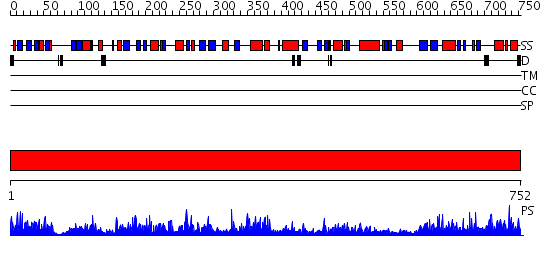
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..752] | 1000.0 | Carbon monoxide (CO) dehydrogenase molybdoprotein, N-domain; Carbon monoxide (CO) dehydrogenase molybdoprotein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)