













| General Information: |
|
| Name(s) found: |
CE41423
[WormBase]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 735 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: |
endodeoxyribonuclease activity, producing 5'-phosphomonoesters
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDLHQHLPSY QEADGGAHGK ETTSHIIYNT SQEYHQINHQ QFDQNNLGEL PIYLNYPLHG 60
61 PNHKKWHLDF QQNNDANEYS QNGNHDFSGA EMSYPIEDFA QNTFAEQYVE DSYQNAGALA 120
121 LTGQNMNISN VHNGVHISTP TENLAENNAN NGLDAIADLV KPYQNVQLES SSKQKVQRPK 180
181 IAHKNSCAHG YKYAVAYQIL KDNVGYEIPV DTVKLCRENH IDTKVLNMVV RRLEELGAIE 240
241 RPLNLHVRFT PEFKFKEIDE FQSTHIEIEK LDQEEKELDE MLEIAQKQLA EVKRNPYAFF 300
301 TSADINLRGN EVLIAKAHDV TVDCMTGMNR VVTNMDAVKI DKMNYLNNRF PKEALTKIDN 360
361 KKYKAAAKII DKNIGQDKVQ LKQKSKDEAV MKILRQFSGT NEASDPSDER PVHQALGINK 420
421 KMAHQTTQNS QLRITTSGNE LKVSIGKKFD EAQINKLQTE KELKKNAQKQ ANAAKMREKR 480
481 AKAKEEQEGS RARKRKNTDE PTLPIKRQDS IDSECALGQL DPYNNRIMFS EMSSPVGCYN 540
541 PDLVPDIERL RQVSDAISDD QVCNQIFNEP QHGLSLSSQY FEGTRSDEPN SVLNEGLNFD 600
601 IPQENLRMLS PRRENFGMLR GREISEENHT LNCFNSQTFD DDIMDFNFHS MQNLPSTSEM 660
661 QFKSSDDNFD GSFSNFEYDN KENEHDEANE SIDFGFLNEE LKMIPDDEHN APIDLSDLSL 720
721 HSHFNENMAD SSLVY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
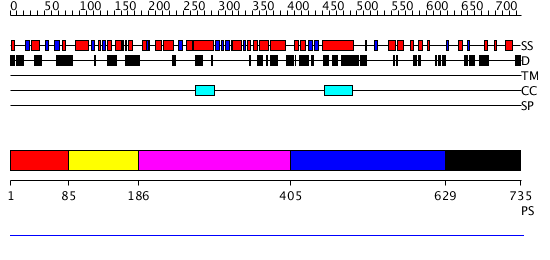
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..84] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [85..185] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [186..404] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [405..628] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [629..735] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)