













| General Information: |
|
| Name(s) found: |
CE41386
[WormBase]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 787 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ribosome
[IEA intracellular [IEA gas vesicle [IEA |
| Biological Process: |
translation
[IEA gas vesicle organization [IEA |
| Molecular Function: |
nucleotide binding
[IEA ATPase activity [IEA ATP binding [IEA structural constituent of ribosome [IEA nucleoside-triphosphatase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MITRVSLLRL QHARKVLTTT ASTSSKPSTL TIGSVSVQLQ DAKNPDFVPA FYGQKNNPPQ 60
61 DLLPHLRWLI QKDKLKQDVF LLGVPGKIRL ELVLRYLEAT NREFEYLPIT RDTTEADIKQ 120
121 RREIRDGTAY YTDLCAVRAA LKGRVLVIDG VERAERNVLP ILNNLLENRE MQLDDGRFLM 180
181 KHDKYDELKV KYDEATLKKM GMERVSENFH VIALGLPVPR FPGNSLDPPF RSRFQCRNIQ 240
241 ELSFHTFLEQ AKLIAPNVSE TNLNDLISVV YAYNSEASTS RPRIGTAVVE NTLKIWNENP 300
301 SYSAGEIVNF SYPAKEVLSD SEKDLINTFK TKFELSTHER TRELKTDGAL HTVQVPADKR 360
361 KIQMKLPTPS HPTSFASTWI PTAKQENLIA DLALAFSKGD FILIGPKGSG KSRILGELSR 420
421 RLNFNYVTMV LHQDMNTREL IQRRHMKENG DTVWEDSILV TAARNGDVCV LDGVEMVHSS 480
481 VIMSLSQLIY HRRFDLPNGN RLVGEKEFLT IMERDGYDAD AYEKVWNKVQ APSRKLASVI 540
541 DQLEAKKKER EWTKHQTSGD LDDGKLIEGV TGEQNIYRRR IDKVPDPGAP QTKPKRLKVC 600
601 LDVSGSMYRF NGYDQRLVKS LEAALMTMTA LDGKTDKVQY DIIGHSGDSP CVSFVKTDHH 660
661 PKNNKERLDT LKRMIAHTQY CVSGDNTVES LQFAIKELAA KKDDFDETVV ILVSDANLER 720
721 YGIQPKELKD AMAKEPNINS FVIFIGSLSD EADQLQRELP VGKAFVLKDT SELPKIMETI 780
781 FSSTIAQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
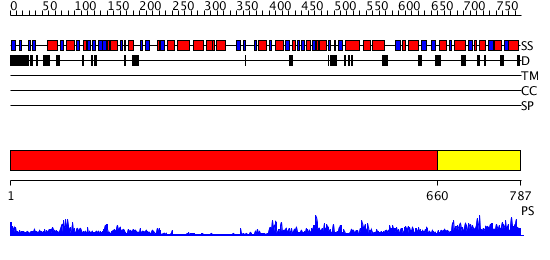
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..659] | 88.69897 | N-terminal, ClpS-binding domain of ClpA, an Hsp100 chaperone; ClpA, an Hsp100 chaperone, AAA+ modules | |
| 2 | View Details | [660..787] | 25.0 | Holliday junction helicase RuvB |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.50 |
Source: Reynolds et al. (2008)