













| General Information: |
|
| Name(s) found: |
UVRA_ECOLI
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Escherichia coli |
| Length: | 940 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDKIEVRGAR THNLKNINLV IPRDKLIVVT GLSGSGKSSL AFDTLYAEGQ RRYVESLSAY 60
61 ARQFLSLMEK PDVDHIEGLS PAISIEQKST SHNPRSTVGT ITEIHDYLRL LFARVGEPRC 120
121 PDHDVPLAAQ TVSQMVDNVL SQPEGKRLML LAPIIKERKG EHTKTLENLA SQGYIRARID 180
181 GEVCDLSDPP KLELQKKHTI EVVVDRFKVR DDLTQRLAES FETALELSGG TAVVADMDDP 240
241 KAEELLFSAN FACPICGYSM RELEPRLFSF NNPAGACPTC DGLGVQQYFD PDRVIQNPEL 300
301 SLAGGAIRGW DRRNFYYFQM LKSLADHYKF DVEAPWGSLS ANVHKVVLYG SGKENIEFKY 360
361 MNDRGDTSIR RHPFEGVLHN MERRYKETES SAVREELAKF ISNRPCASCE GTRLRREARH 420
421 VYVENTPLPA ISDMSIGHAM EFFNNLKLAG QRAKIAEKIL KEIGDRLKFL VNVGLNYLTL 480
481 SRSAETLSGG EAQRIRLASQ IGAGLVGVMY VLDEPSIGLH QRDNERLLGT LIHLRDLGNT 540
541 VIVVEHDEDA IRAADHVIDI GPGAGVHGGE VVAEGPLEAI MAVPESLTGQ YMSGKRKIEV 600
601 PKKRVPANPE KVLKLTGARG NNLKDVTLTL PVGLFTCITG VSGSGKSTLI NDTLFPIAQR 660
661 QLNGATIAEP APYRDIQGLE HFDKVIDIDQ SPIGRTPRSN PATYTGVFTP VRELFAGVPE 720
721 SRARGYTPGR FSFNVRGGRC EACQGDGVIK VEMHFLPDIY VPCDQCKGKR YNRETLEIKY 780
781 KGKTIHEVLD MTIEEAREFF DAVPALARKL QTLMDVGLTY IRLGQSATTL SGGEAQRVKL 840
841 ARELSKRGTG QTLYILDEPT TGLHFADIQQ LLDVLHKLRD QGNTIVVIEH NLDVIKTADW 900
901 IVDLGPEGGS GGGEILVSGT PETVAECEAS HTARFLKPML |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
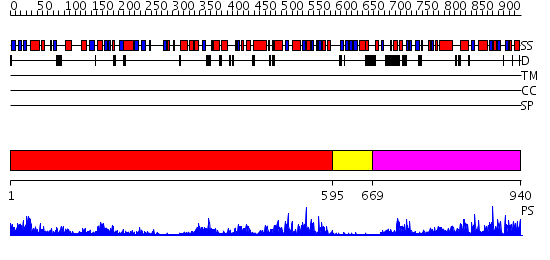
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..594] | 32.0 | RNase-L Inhibitor | |
| 2 | View Details | [595..668] | 2.67 | MJ0796 | |
| 3 | View Details | [669..940] | 55.0 | No description for 2d62A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)