













| General Information: |
|
| Name(s) found: |
HDG3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 6 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 725 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA][ISS]
|
| Biological Process: |
regulation of transcription, DNA-dependent
[ISS]
cotyledon development [IGI] |
| Molecular Function: |
DNA binding
[ISS]
transcription factor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQSNMVPVA NNGDNNNDNE NNNNNNNNGG TDNTNAGNDS GDQDFDSGNT SSGNHGEGLG 60
61 NNQAPRHKKK KYNRHTQLQI SEMEAFFREC PHPDDKQRYD LSAQLGLDPV QIKFWFQNKR 120
121 TQNKNQQERF ENSELRNLNN HLRSENQRLR EAIHQALCPK CGGQTAIGEM TFEEHHLRIL 180
181 NARLTEEIKQ LSVTAEKISR LTGIPVRSHP RVSPPNPPPN FEFGMGSKGN VGNHSRETTG 240
241 PADANTKPII MELAFGAMEE LLVMAQVAEP LWMGGFNGTS LALNLDEYEK TFRTGLGPRL 300
301 GGFRTEASRE TALVAMCPTG IVEMLMQENL WSTMFAGIVG RARTHEQIMA DAAGNFNGNL 360
361 QIMSAEYQVL SPLVTTRESY FVRYCKQQGE GLWAVVDISI DHLLPNINLK CRRRPSGCLI 420
421 QEMHSGYSKV TWVEHVEVDD AGSYSIFEKL ICTGQAFAAN RWVGTLVRQC ERISSILSTD 480
481 FQSVDSGDHI TLTNHGKMSM LKIAERIART FFAGMTNATG STIFSGVEGE DIRVMTMKSV 540
541 NDPGKPPGVI ICAATSFWLP APPNTVFDFL REATHRHNWD VLCNGEMMHK IAEITNGIDK 600
601 RNCASLLRHG HTSKSKMMIV QETSTDPTAS FVLYAPVDMT SMDITLHGGG DPDFVVILPS 660
661 GFAIFPDGTG KPGGKEGGSL LTISFQMLVE SGPEARLSVS SVATTENLIR TTVRRIKDLF 720
721 PCQTA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
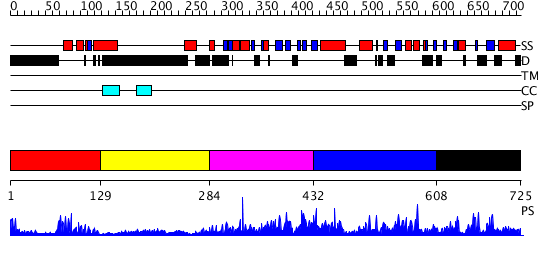
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..128] | 14.39794 | Homeobox protein hox-b1 | |
| 2 | View Details | [129..283] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [284..431] | 8.30103 | Smad1 | |
| 4 | View Details | [432..607] | 1.059965 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [608..725] | 3.058984 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)