













| General Information: |
|
| Name(s) found: |
APRR7_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 727 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
mitochondrion [IDA] |
| Biological Process: |
response to temperature stimulus
[IGI]
red or far-red light signaling pathway [IMP] circadian rhythm [IMP][TAS] |
| Molecular Function: |
DNA binding
[IPI]
transcription regulator activity [TAS] transcription repressor activity [IDA] two-component response regulator activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNANEEGEGS RYPITDRKTG ETKFDRVESR TEKHSEEEKT NGITMDVRNG SSGGLQIPLS 60
61 QQTAATVCWE RFLHVRTIRV LLVENDDCTR YIVTALLRNC SYEVVEASNG IQAWKVLEDL 120
121 NNHIDIVLTE VIMPYLSGIG LLCKILNHKS RRNIPVIMMS SHDSMGLVFK CLSKGAVDFL 180
181 VKPIRKNELK ILWQHVWRRC QSSSGSGSES GTHQTQKSVK SKSIKKSDQD SGSSDENENG 240
241 SIGLNASDGS SDGSGAQSSW TKKAVDVDDS PRAVSLWDRV DSTCAQVVHS NPEFPSNQLV 300
301 APPAEKETQE HDDKFEDVTM GRDLEISIRR NCDLALEPKD EPLSKTTGIM RQDNSFEKSS 360
361 SKWKMKVGKG PLDLSSESPS SKQMHEDGGS SFKAMSSHLQ DNREPEAPNT HLKTLDTNEA 420
421 SVKISEELMH VEHSSKRHRG TKDDGTLVRD DRNVLRRSEG SAFSRYNPAS NANKISGGNL 480
481 GSTSLQDNNS QDLIKKTEAA YDCHSNMNES LPHNHRSHVG SNNFDMSSTT ENNAFTKPGA 540
541 PKVSSAGSSS VKHSSFQPLP CDHHNNHASY NLVHVAERKK LPPQCGSSNV YNETIEGNNN 600
601 TVNYSVNGSV SGSGHGSNGP YGSSNGMNAG GMNMGSDNGA GKNGNGDGSG SGSGSGSGNL 660
661 ADENKISQRE AALTKFRQKR KERCFRKKVR YQSRKKLAEQ RPRVRGQFVR KTAAATDDND 720
721 IKNIEDS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
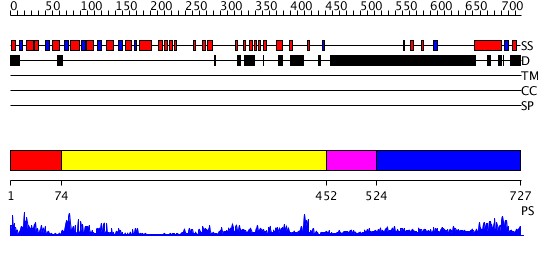
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..73] | 18.0 | No description for 2ayxA was found. | |
| 2 | View Details | [74..451] | 62.154902 | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | |
| 3 | View Details | [452..523] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [524..727] | 8.044979 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)