













| General Information: |
|
| Name(s) found: |
MDAR2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 435 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA]
|
| Biological Process: |
response to zinc ion
[IEP]
response to salt stress [IEP] response to cadmium ion [IEP] |
| Molecular Function: |
monodehydroascorbate reductase (NADH) activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEEKSFKYV IVGGGVAAGY AAREFFNQGV KPGELAIISR EQVPPYERPA LSKGYIHLEN 60
61 KATLPNFYVA AGIGGERQFP QWYKEKGIEL ILGTEIVKAD LAAKTLVSGT GQVFKYQTLL 120
121 AATGSSVIRL SDFGVPGADA KNIFYLRELE DADYLAYAME TKEKGKAVVV GGGYIGLELG 180
181 AALKANNLDV TMVYPEPWCM PRLFTAGIAS FYEGYYANKG INIVKGTVAS GFTTNSNGEV 240
241 TEVKLKDGRT LEADIVIVGV GGRPIISLFK DQVEEEKGGL KTDGFFKTSL PDVYAIGDVA 300
301 TFPMKLYNEM RRVEHVDHAR KSAEQAVKAI KAAEEGNSIP EYDYLPYFYS RAFDLSWQFY 360
361 GDNVGESVLF GDNDPESPKP KFGSYWIKER KVVGAFLEGG SPEENNAIAK LARAQPSVES 420
421 LEVLSKEGLS FATNI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
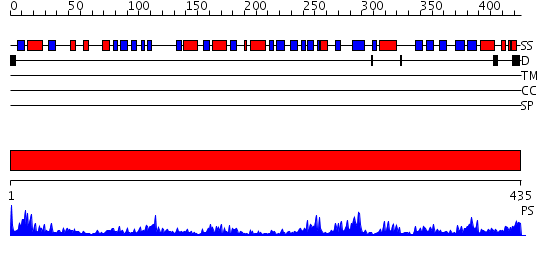
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..435] | 65.69897 | Crystal Structure of Putidaredoxin Reductase from Pseudomonas putida |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)