













| General Information: |
|
| Name(s) found: |
gi|151944130
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae YJM789 |
| Length: | 1557 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPEIYGPQPL KPLNTVMRHG FEEQYQSDQL LQSLANDFIF YFDDKRHKTN GNPIPEEDKQ 60
61 RDVNRYYQPI TDWKIMKDRQ KTVSAALLLC LNLGVDPPDV MKTHPCARVE AWVDPLNFQD 120
121 SKKAIEQIGK NLQAQYETLS LRTRYKQSLD PCVEDVKRFC NSLRRTSKED RILFHYNGHG 180
181 VPKPTKSGEI WVFNRGYTQY IPVSLYDLQT WLGAPCIFVY DCNSAENILI NFQKFVQKRI 240
241 KDDEEGNHDV AAPSPTSAYQ DCFQLASCTS DELLLMSPEL PADLFSCCLT CPIEISIRIF 300
301 LMQSPLKDSK YKIFFENSTS NQPFGDSKNS FKSKIPNVNI PGMLSDRRTP LGELNWIFTA 360
361 ITDTIAWTSL PRPLFKKLFR HDLMIAALFR NFLLAKRIMP WYNCHPVSDP ELPDSITTHP 420
421 MWKSWDLAMD EVLTKIVIDL KNAPPATALE SQMILQQQET LQNGGSSKSN AQDTKAGSIQ 480
481 TQSRFAVANL STMSLVNNPA LQSRKSISLQ SSQQQLQQQQ QQQQQFTGFF EQNLTAFELW 540
541 LKYASNVRHP PEQLPIVLQV LLSQVHRIRA LVLLSRFLDL GPWAVYLSLS IGIFPYVLKL 600
601 LQSPAPELKP ILVFIWARIM SIDYKNTQSE LIKEKGYMYF VTVLVPDWGV NGMSATNGSA 660
661 MINSGNPLTM TASQNINGPS SRYYERQQGN RTSNLGHNNL PFYHSNDTTD EQKAMAVFVL 720
721 ASFVRNFPLG QKNCFSLELV NKLCFYIDNS EIPLLRQWCV ILLGLLFADN PLNRFVCMNT 780
781 GAVEILLKSL KDPVPEVRTA SIFALKHFIS GFQDAEVILR LQQEFEEQYQ QLHSQLQHLQ 840
841 NQSHLQQQQS QQQQQHLEQQ QMKIEKQIRH CQVMQNQLEV IDLRKLKRQE IGNLISILPL 900
901 INDGSSLVRK ELVVYFSHIV SRYSNFFIVV VFNDLLEEIK LLEKSDINTR NTSDKYSVSQ 960
961 GSIFYTVWKS LLILAEDPFL ENKELSKQVI DYILLELSAH KELGGPFAVM EKFLLKRSSK1020
1021 AHQTGKFGFN SSQVQFVKSS LRSFSPNERV DNNAFKKEQQ QHDPKISHPM RTSLAKLFQS1080
1081 LGFSESNSDS DTQSSNTSMK SHTSKKGPSG LYLLNGNNNI YPTAETPRFR KHTEPLQLPL1140
1141 NSSFLDYSRE YFQEPQMKKQ EADEPGSVEY NARLWRRNRN ETIIQETQGE KKLSIYGNWS1200
1201 KKLISLNNKS QPKLMKFAQF EDQLITADDR STITVFDWEK GKTLSKFSNG TPFGTKVTDL1260
1261 KLINEDDSAL LLTGSSDGVI KIYRDYQDVD TFKIVSAWRG LTDMLLTPRS TGLLTEWLQI1320
1321 RGSLLTTGDV KVIRVWDAHT ETVEVDIPAK TSSLITSLTA DQLAGNIFVA GFADGSLRVY1380
1381 DRRLDPRDSM IRRWRAGNDK QGVWINNVHL QRGGYRELVS GATNGVVELW DIRSEDPVES1440
1441 FVDQNVTSQY GSQQKPTTMT CMQVHEHAPI IATGTKQIKI WTTSGDLLNS FKNSHNNGVT1500
1501 STLAATGIPK SLSYSSTSDA FLSSMAFHPH RMMIAATNSH DSIVNIYKCE DERIDYF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
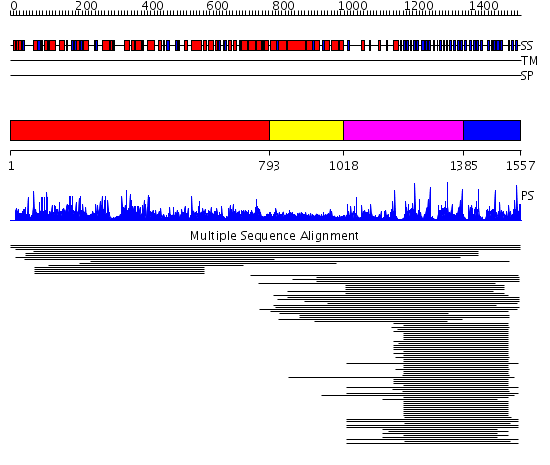
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..792] | 1.52 | Karyopherin alpha | |
| 2 | View Details | [793..1017] | 3.082997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [1018..1384] | 17.69897 | Arp2/3 complex 41 kDa subunit ARPC1 | |
| 4 | View Details | [1385..1557] | 75.522879 | beta1-subunit of the signal-transducing G protein heterotrimer | |
| 5 | View Details | [414..948] | 1.26 | Crystal structure of the exportin Cse1p complexed with its cargo ( Kap60p) and RanGTP | |
| 6 | View Details | [949..1029] | 1.42 | Mouse Importin alpha- phosphorylated SV40 CN peptide complex | |
| 7 | View Details | [1030..1136] | 19.69897 | Arp2/3 complex 41 kDa subunit ARPC1 | |
| 8 | View Details | [1137..1557] | 76.0 | No description for 2ovpB was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.72 |
Source: Reynolds et al. (2008)