













| General Information: |
|
| Name(s) found: |
gi|151940917
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae YJM789 |
| Length: | 691 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNRFWNTKKF SLTNADGLCA TLNEISQNDE VLVVQPSVLP VLNSLLTFQD LTQSTPVRKI 60
61 TLLDDQLSDD LPSALGSVPQ MDLIFLIDVR TSLRLPPQLL DAAQKHNLSS LHIIYCRWKP 120
121 SFQNTLEDTE QWQKDGFDLN SKKTHFPNVI ESQLKELSNE YTLYPWDLLP FPQIDENVLL 180
181 THSLYNMENV NMYYPNLRSL QSATESILVD DMVNSLQSLI FETNSIITNV VSIGNLSKRC 240
241 SHLLKKRIDE HQTENDLFIK GTLYGERTNC GLEMDLIILE RNTDPITPLL TQLTYAGILD 300
301 DLYEFNSGIK IKEKDMNFNY KEDKIWNDLK FLNFGSIGPQ LNKLAKELQT QYDTRHKAES 360
361 VHEIKEFVDS LGSLQQRQAF LKNHTTLSSD VLKVVETEEY GSFNKILELE LEILMGNTLN 420
421 NDIEDIILEL QYQYEVDQKK ILRLICLLSL CKNSLREKDY EYLRTFMIDS WGIEKCFQLE 480
481 SLAELGFFTS KTGKTDLHIT TSKSTRLQKE YRYISQWFNT VPIEDEHAAD KITNENDDFS 540
541 EATFAYSGVV PLTMRLVQML YDRSILFHNY SSQQPFILSR EPRVSQTEDL IEQLYGDSHA 600
601 IEESIWVPGT ITKKINASIK SNNRRSIDGS NGTFHAAEDI ALVVFLGGVT MGEIAIMKHL 660
661 QKILGKKGIN KRFIIIADGL INGTRIMNSI S |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
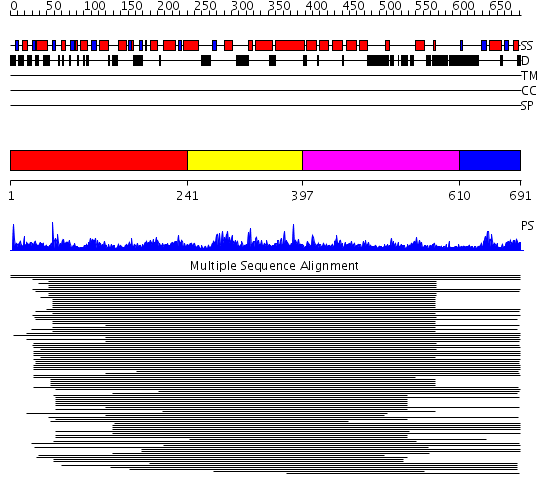
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..240] | 166.9897 | Neuronal Sec1, NSec1 | |
| 2 | View Details | [241..396] | 166.9897 | Neuronal Sec1, NSec1 | |
| 3 | View Details | [397..609] | 166.9897 | Neuronal Sec1, NSec1 | |
| 4 | View Details | [610..691] | 1.036988 | View MSA. Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
|||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.62 |
Source: Reynolds et al. (2008)