













| General Information: |
|
| Name(s) found: |
gi|151945985
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae YJM789 |
| Length: | 732 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTQIRSPQG LPYPIQIDKL IPSVGSYLHE GDRLLVYKFW YLVERASDTG DDDNEHDVSP 60
61 GGSAGSNGVS PPTKQLRESI EFFESPYEGD LISWNVDVGD EVATANQVIC EIKRPCNHDI 120
121 VYGGLCTQCG KEVSADAFDG VPLDVVGDVD LQISETEAIR TGKALKEHLR RDKKLILVVD 180
181 LDQTIIHCGV DPTIAEWKND PNNPNFETLR DVKSFTLDEE LVLPLMYMND DGSMLRPPPV 240
241 RKCWYYVKVR PGLKEFFAKV APLFEMHIYT MATRAYALQI AKIVDPTGEL FGDRILSRDE 300
301 NGSLTTKSLA KLFPTDQSMV VVIDDRGDVW NWCPNLIKVV PYNFFVGVGD INSNFLPKQS 360
361 TGMLQLGRKT RQKSQESQEL LTDIMDNEKK LQEKIDKEVK RQEEKLNHQL ATAEEPPANE 420
421 SKEELTKKLE YSASLEVQQQ NRPLAKLQKH LHDQKLLVDD DDELYYLMGT LSNIHKTYYD 480
481 MLSQQNEPEP NLMEIIPSLK QKVFQNCYFV FSGLIPLGTD IQRSDIVIWT STFGATSTPD 540
541 IDYLTTHLIT KNPSTYKARL AKKFNPQIKI VHPDWIFECL VNWKKVDEKP YTLIVDSPIS 600
601 DEELQNFQTQ LQKRQEYLEE TQEQQHMLTS QENLNLFAAG TSWLNNDDDE DIPDTASDDD 660
661 EDDDHDDESD DENNSEGIDR KRSIEDNHDD TSQKKTKAEP SQDGPVQHKG EGDDNEDSDS 720
721 QLEEELMDML DD |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
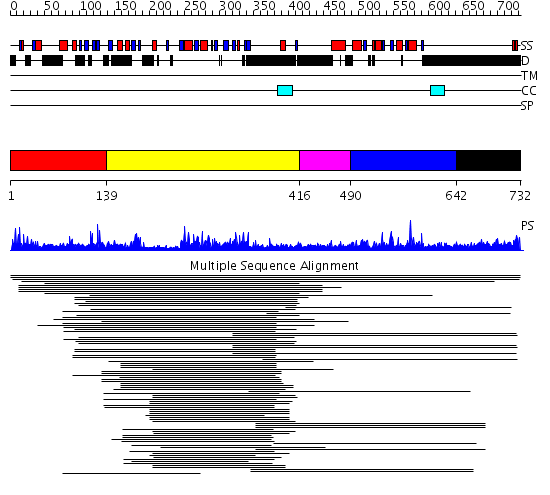
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..138] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [139..415] | 9.886057 | NLI interacting factor-like phosphatase No confident structure predictions are available. | |
| 3 | View Details | [416..489] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [490..641] | 12.522879 | Breast cancer associated protein, BRCA1 | |
| 5 | View Details | [642..732] | 1.045991 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)