













| General Information: |
|
| Name(s) found: |
gcy-37 /
CE37494
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 708 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
cGMP biosynthetic process
[IEA embryonic development ending in birth or egg hatching [IMP cyclic nucleotide biosynthetic process [IEA |
| Molecular Function: |
phosphorus-oxygen lyase activity
[IEA guanylate cyclase activity [IEA heme binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIGWTHVCVS ALILRKYGPE VLEEILRKAG YQEDIKFDIQ CYYDDTETMR IFRVAATVLG 60
61 LSVDDMWEMY GEFLITHACE TGWQKMLFCM ANNLQEFLDN LNSMHYFIDQ IAFKSEMKGP 120
121 TFQCEPFGES GLKLHYFSFR QGLFPIVKGL VRKTARTLFE MDVKVCMLER NQERRKSGMV 180
181 EHVIFSVEPD DNHRKGKRLF HKFRNTKTTE NAPSFTLSST ILVGLRDFKN IFPYHVCFNK 240
241 QMIIEHIGIY LLREYGLENK KTLKVSDLMQ LVQPSDIQLT YKNVLSYLNT LFIFQLKHHS 300
301 KRNEVQEGSS EAFQQPLVLK GEMMPINDGN SIIFICSPHV TTVRDILNLK LYISDMPMHD 360
361 ATRDLVMLNQ SRICQMELNK KLEETMKKMK KMTEELEVKK SQTDRLLFEF VPPVIAEALR 420
421 AAKTVPAQEF SDCSVIFTDI PDFFTISVNC SPTEIITVVT DLFHRFDRII EKHKGYKVLS 480
481 LMDSYLIVGG VPNANQYHCE DSLNLALGLL FEAKQVVVPK LERSVRLRIG VHCGPVVAGI 540
541 VSQQKPRFCV LGNTVNVTKS ICSHSSPGKV LVSNAVRTMV TKHLKSIFVF NANGYLELQS 600
601 GKVLTHFLEK NEKCSVWDIV DRDKATNDSI DGYRELHSDN GTEEWQEATV AAYRVISVVD 660
661 ALENKQSRTR KALTRLRSVK RKFRTIQSND SGVSVSEPNV ESAVCSIM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
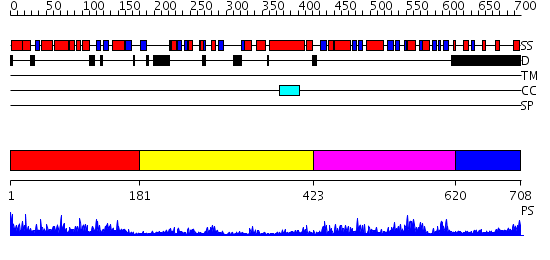
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..180] | 41.522879 | No description for 2o09A was found. | |
| 2 | View Details | [181..422] | 5.30103 | No description for 2evaA was found. | |
| 3 | View Details | [423..619] | 49.69897 | Crystal structure of a mutant form of the mycobacterial adenylyl cyclase Rv1625c | |
| 4 | View Details | [620..708] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)