













| General Information: |
|
| Name(s) found: |
ARFG_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1164 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
phototropism
[IMP]
leaf development [IGI] regulation of transcription, DNA-dependent [TAS] blue light signaling pathway [IMP] gravitropism [TAS] regulation of growth [TAS] lateral root primordium development [IGI] response to ethylene stimulus [IGI] response to auxin stimulus [IMP] lateral root development [IGI] |
| Molecular Function: |
DNA binding
[IDA]
transcription factor activity [ISS] transcription regulator activity [ISS] transcription activator activity [IMP] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKAPSSNGVS PNPVEGERRN INSELWHACA GPLISLPPAG SLVVYFPQGH SEQVAASMQK 60
61 QTDFIPSYPN LPSKLICMLH NVTLNADPET DEVYAQMTLQ PVNKYDRDAL LASDMGLKLN 120
121 RQPNEFFCKT LTASDTSTHG GFSVPRRAAE KIFPALDFSM QPPCQELVAK DIHDNTWTFR 180
181 HIYRGQPKRH LLTTGWSVFV STKRLFAGDS VLFIRDGKAQ LLLGIRRANR QQPALSSSVI 240
241 SSDSMHIGVL AAAAHANANN SPFTIFYNPR AAPAEFVVPL AKYTKAMYAQ VSLGMRFRMI 300
301 FETEECGVRR YMGTVTGISD LDPVRWKNSQ WRNLQIGWDE SAAGDRPSRV SVWDIEPVLT 360
361 PFYICPPPFF RPRFSGQPGM PDDETDMESA LKRAMPWLDN SLEMKDPSST IFPGLSLVQW 420
421 MNMQQQNGQL PSAAAQPGFF PSMLSPTAAL HNNLGGTDDP SKLLSFQTPH GGISSSNLQF 480
481 NKQNQQAPMS QLPQPPTTLS QQQQLQQLLH SSLNHQQQQS QSQQQQQQQQ LLQQQQQLQS 540
541 QQHSNNNQSQ SQQQQQLLQQ QQQQQLQQQH QQPLQQQTQQ QQLRTQPLQS HSHPQPQQLQ 600
601 QHKLQQLQVP QNQLYNGQQA AQQHQSQQAS THHLQPQLVS GSMASSVITP PSSSLNQSFQ 660
661 QQQQQSKQLQ QAHHHLGAST SQSSVIETSK SSSNLMSAPP QETQFSRQVE QQQPPGLNGQ 720
721 NQQTLLQQKA HQAQAQQIFQ QSLLEQPHIQ FQLLQRLQQQ QQQQFLSPQS QLPHHQLQSQ 780
781 QLQQLPTLSQ GHQFPSSCTN NGLSTLQPPQ MLVSRPQEKQ NPPVGGGVKA YSGITDGGDA 840
841 PSSSTSPSTN NCQISSSGFL NRSQSGPAIL IPDAAIDMSG NLVQDLYSKS DMRLKQELVG 900
901 QQKSKASLTD HQLEASASGT SYGLDGGENN RQQNFLAPTF GLDGDSRNSL LGGANVDNGF 960
961 VPDTLLSRGY DSQKDLQNML SNYGGVTNDI GTEMSTSAVR TQSFGVPNVP AISNDLAVND1020
1021 AGVLGGGLWP AQTQRMRTYT KVQKRGSVGR SIDVNRYRGY DELRHDLARM FGIEGQLEDP1080
1081 QTSDWKLVYV DHENDILLVG DDPWEEFVNC VQSIKILSSA EVQQMSLDGN FAGVPVTNQA1140
1141 CSGGDSGNAW RGHYDDNSAT SFNR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
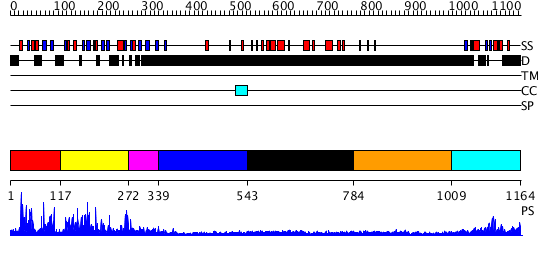
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..116] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [117..271] | 14.09691 | Solution Structure of the B3 DNA-Binding Domain of RAV1 | |
| 3 | View Details | [272..338] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [339..542] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [543..783] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [784..1008] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1009..1164] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.71 |
Source: Reynolds et al. (2008)