













| General Information: |
|
| Name(s) found: |
spd-1 /
CE37061
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 529 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA spindle midzone [IDA microtubule [IDA |
| Biological Process: |
negative regulation of multicellular organism growth
[IMP regulation of multicellular organism growth [IMP hermaphrodite genitalia development [IMP embryonic development ending in birth or egg hatching [IMP post-embryonic body morphogenesis [IMP positive regulation of growth rate [IMP gamete generation [IMP mitotic spindle stabilization [IMP positive regulation of locomotion [IMP gonad development [IMP cytokinesis, completion of separation [IMP locomotory behavior [IMP morphogenesis of an epithelium [IMP oviposition [IMP |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRRHSVVDA YSTVEGKIRD TMRELSALWD EVDMSEAMRL KRVDNAFTHI TLLCDDMLSG 60
61 EKEMIHNLKV SIREDMQNVK KMRLELEMED FQRPAEIKDG SIALMRHLQS EVKSLETEFQ 120
121 TRHEDQRVLI EKICNLKKRL ESDFEFEYEI HALFPVKAFE KYSASCSEME ELLKHRYHQI 180
181 ERLQTDISQW KSMAQNVADY IGGDDDLKNL LEKNIDSPEF VFSERLLDSL QSYHSELEPL 240
241 YKHWLEDIEF RWTEKHQQLQ ELWEKCLVSE ADRHYSPIFE PTKNSEQTLK RMEDECDRLE 300
301 KKYEACRIVF ELVDKWRAVW TEKLTIDEKR KQPNYYKTTN VLPDNKRERE LLNQMPVLER 360
361 KIDSAHRKYG NEHDGEQISI QGMAPLEYVR FVQEEHKREL KFELQLKKEE KTRMQSPTPT 420
421 RTPRTQKRAM FRTPMSTAKM EPVAKKLNFD TDLPPCMSPA TSEMISFITP TRRQAVGPKT 480
481 SSPKDTQPST SSSSRTTTPM SKRAMTPSSI ASSTPSSAKK VLTRRNQFL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
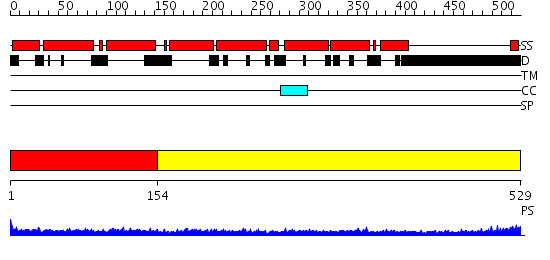
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..153] | 4.69897 | No description for 2i1jA was found. | |
| 2 | View Details | [154..529] | 15.522879 | Heavy meromyosin subfragment |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)