













| General Information: |
|
| Name(s) found: |
dab-1 /
CE36393
[WormBase]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 474 amino acids |
Gene Ontology: |
|
| Cellular Component: |
AP-1 adaptor complex
[IDA vesicle [IDA AP-2 adaptor complex [IDA Golgi-associated vesicle [IDA |
| Biological Process: |
molting cycle, protein-based cuticle
[IMP vesicle-mediated transport [IEP locomotory behavior [IMP positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP oviposition [IMP protein secretion [IMP cell migration [IMP positive regulation of locomotion [IMP |
| Molecular Function: |
clathrin heavy chain binding
[IDA protein domain specific binding [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKKTQLGRSF SKSFKQKKAR ASNASSDPFR FQNNGISYKG KLIGEQDVDK ARGDAMCAEA 60
61 MRTAKSIIKA AGAHKTRITL QINIDGIKVL DEKSGAVLHN FPVSRISFIA RDSSDARAFG 120
121 LVYGEPGGKY KFYGIKTAQA ADQAVLAIRD MFQVVFEMKK KQIEQVKQQQ IQDGGAEISS 180
181 KKEGGVAVAD LLDLESELQQ IERGVQQLST VPTNCDAFGA SPFGDPFVDS FNSTATSNGT 240
241 ANMSGTQVPF GGLQLPQVQQ MQMPMVQIPQ QSHQNWPTSG AGSFDAWGQQ QQQQQMHHAH 300
301 STPAFGTNGF SDTNPFASAF NTQARPPPLP TVAPTQYRDP FSVHSSAIPN SNIDWTGTGT 360
361 TKENMAPSTN IQQQQSSLHH ASTFANFGDN KAETWSEKKV TSLEEAFTKL VDMDALVGGQ 420
421 GIKETKKNPF EHILNPPKAS LNSMSTTCSA AQMAATQQHT TSSHADPFGD DFFR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
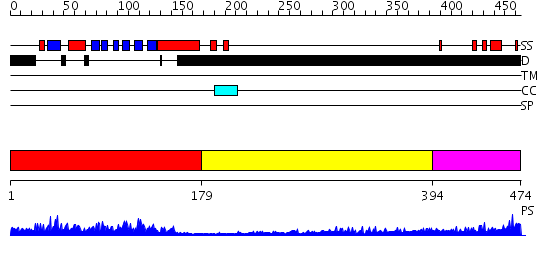
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..178] | 51.0 | Crystal structure of the phosphotyrosine binding domain (PTB) of mouse Disabled 1 (Dab1) | |
| 2 | View Details | [179..393] | 4.193997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [394..474] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)