













| General Information: |
|
| Name(s) found: |
prx-5 /
CE36377
[WormBase]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 500 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
protein import into peroxisome matrix
[IMP nematode larval development [IMP positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP response to stress [IEA growth [IMP |
| Molecular Function: |
peroxisome matrix targeting signal-1 binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKGVVEGQCG QQNALVGLAN TFGTSNQRVA PSNASLLPSS SMGEQMANEF LRQQARTMAP 60
61 TSFSMKSMQN NLPQASASSS LAANWTKEFQ PRQNQLASQW SQQYTSSAPS MESAWRQVQA 120
121 PSMTSTSSHQ PITDAGMWSS EYLDTVDTSL TKSSGTQNWA DDFMEQQDNY GMENTWKDAQ 180
181 AFEQRWEEIK RDMEKDESLQ SPENYVYQEA NPFTTMSDPL MEGDNLMRNG DIGNAMLAYE 240
241 AAVQKDPQDA RAWCKLGLAH AENEKDQLAM QAFQKCLQID AGNKEALLGL SVSQANEGME 300
301 NEALHQLDKW MSSYLGSNST QVTTTPPLYS SFLDSDTFNR VEARFLDAAR QQGATPDPDL 360
361 QNALGVLYNL NRNFARAVDS LKLAISKNPT DARLWNRLGA TLANGDHTAE AISAYREALK 420
421 LYPTYVRARY NLGISCMQLS SYDEALKHFL SALELQKGGN DASGIWTTMR SAAIRTSNVP 480
481 DNLLRAVERR DLAAVKASLV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
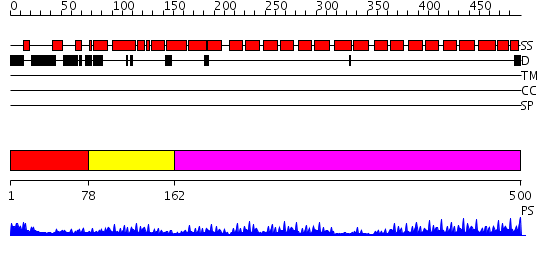
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..77] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [78..161] | 22.0 | No description for 2q7fA was found. | |
| 3 | View Details | [162..500] | 29.39794 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)