













| General Information: |
|
| Name(s) found: |
AB40G_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1423 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA][IEP]
|
| Biological Process: |
response to biotic stimulus
[TAS]
drug transmembrane transport [ISS] lead ion transport [IMP] response to ozone [IEP] response to jasmonic acid stimulus [TAS] terpenoid transport [IDA] response to salicylic acid stimulus [TAS] response to ethylene stimulus [TAS] |
| Molecular Function: |
ATPase activity, coupled to transmembrane movement of substances
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEGTSFHQAS NSMRRNSSVW KKDSGREIFS RSSREEDDEE ALRWAALEKL PTFDRLRKGI 60
61 LTASHAGGPI NEIDIQKLGF QDTKKLLERL IKVGDDEHEK LLWKLKKRID RVGIDLPTIE 120
121 VRFDHLKVEA EVHVGGRALP TFVNFISNFA DKFLNTLHLV PNRKKKFTIL NDVSGIVKPG 180
181 RMALLLGPPS SGKTTLLLAL AGKLDQELKQ TGRVTYNGHG MNEFVPQRTA AYIGQNDVHI 240
241 GEMTVRETFA YAARFQGVGS RYDMLTELAR REKEANIKPD PDIDIFMKAM STAGEKTNVM 300
301 TDYILKILGL EVCADTMVGD DMLRGISGGQ KKRVTTGEML VGPSRALFMD EISTGLDSST 360
361 TYQIVNSLRN YVHIFNGTAL ISLLQPAPET FNLFDDIILI AEGEIIYEGP RDHVVEFFET 420
421 MGFKCPPRKG VADFLQEVTS KKDQMQYWAR RDEPYRFIRV REFAEAFQSF HVGRRIGDEL 480
481 ALPFDKTKSH PAALTTKKYG VGIKELVKTS FSREYLLMKR NSFVYYFKFG QLLVMAFLTM 540
541 TLFFRTEMQK KTEVDGSLYT GALFFILMML MFNGMSELSM TIAKLPVFYK QRDLLFYPAW 600
601 VYSLPPWLLK IPISFMEAAL TTFITYYVIG FDPNVGRLFK QYILLVLMNQ MASALFKMVA 660
661 ALGRNMIVAN TFGAFAMLVF FALGGVVLSR DDIKKWWIWG YWISPIMYGQ NAILANEFFG 720
721 HSWSRAVENS SETLGVTFLK SRGFLPHAYW YWIGTGALLG FVVLFNFGFT LALTFLNSLG 780
781 KPQAVIAEEP ASDETELQSA RSEGVVEAGA NKKRGMVLPF EPHSITFDNV VYSVDMPQEM 840
841 IEQGTQEDRL VLLKGVNGAF RPGVLTALMG VSGAGKTTLM DVLAGRKTGG YIDGNITISG 900
901 YPKNQQTFAR ISGYCEQTDI HSPHVTVYES LVYSAWLRLP KEVDKNKRKI FIEEVMELVE 960
961 LTPLRQALVG LPGESGLSTE QRKRLTIAVE LVANPSIIFM DEPTSGLDAR AAAIVMRTVR1020
1021 NTVDTGRTVV CTIHQPSIDI FEAFDELFLL KRGGEEIYVG PLGHESTHLI NYFESIQGIN1080
1081 KITEGYNPAT WMLEVSTTSQ EAALGVDFAQ VYKNSELYKR NKELIKELSQ PAPGSKDLYF1140
1141 PTQYSQSFLT QCMASLWKQH WSYWRNPPYT AVRFLFTIGI ALMFGTMFWD LGGKTKTRQD1200
1201 LSNAMGSMYT AVLFLGLQNA ASVQPVVNVE RTVFYREQAA GMYSAMPYAF AQVFIEIPYV1260
1261 LVQAIVYGLI VYAMIGFEWT AVKFFWYLFF MYGSFLTFTF YGMMAVAMTP NHHIASVVSS1320
1321 AFYGIWNLFS GFLIPRPSMP VWWEWYYWLC PVAWTLYGLI ASQFGDITEP MADSNMSVKQ1380
1381 FIREFYGYRE GFLGVVAAMN VIFPLLFAVI FAIGIKSFNF QKR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
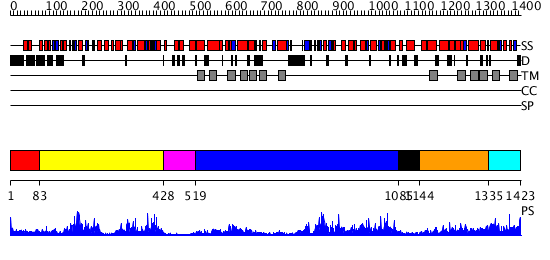
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [83..427] | 42.0 | No description for 2cbzA was found. | |
| 3 | View Details | [428..518] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [519..1084] | 86.522879 | No description for 2hydA was found. | |
| 5 | View Details | [1085..1143] | 62.221849 | Crystal structure of E. coli MalK in the nucleotide-free form | |
| 6 | View Details | [1144..1334] | 2.305992 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1335..1423] | N/A | No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|