













| General Information: |
|
| Name(s) found: |
CSE_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 332 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
response to oxidative stress
[IMP]
response to zinc ion [IEP] response to cadmium ion [IMP] response to hydrogen peroxide [IEP][IMP] |
| Molecular Function: |
lysophospholipase activity
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPSEAESSAN SAPATPPPPP NFWGTMPEEE YYTSQGVRNS KSYFETPNGK LFTQSFLPLD 60
61 GEIKGTVYMS HGYGSDTSWM FQKICMSFSS WGYAVFAADL LGHGRSDGIR CYMGDMEKVA 120
121 ATSLAFFKHV RCSDPYKDLP AFLFGESMGG LVTLLMYFQS EPETWTGLMF SAPLFVIPED 180
181 MKPSKAHLFA YGLLFGLADT WAAMPDNKMV GKAIKDPEKL KIIASNPQRY TGKPRVGTMR 240
241 ELLRKTQYVQ ENFGKVTIPV FTAHGTADGV TCPTSSKLLY EKASSADKTL KIYEGMYHSL 300
301 IQGEPDENAE IVLKDMREWI DEKVKKYGSK TA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
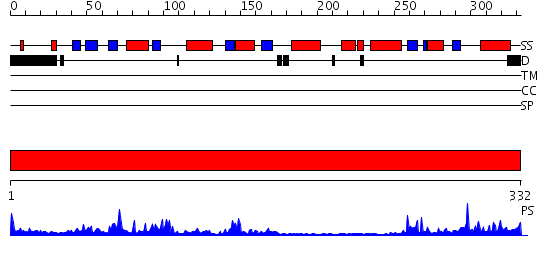
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..332] | 36.0 | Epoxide hydrolase, N-terminal domain; Mammalian epoxide hydrolase, C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.60 |
Source: Reynolds et al. (2008)