













| General Information: |
|
| Name(s) found: |
VIP4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 625 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA]
|
| Biological Process: |
negative regulation of flower development
[IMP]
vernalization response [IMP] |
| Molecular Function: |
protein binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVKGEKRSEM MLNLFGDNSE EEEIESEHEC NRRQPNYASD EAEGGVEPEG EGEAEVEVHG 60
61 EAEAESDGEQ GDVELDPGES EGEREQSSQE ADPQEESEAR DSDSDNKEEE HGGRVAKKRR 120
121 QEVVESGSER SGEKHYESED EEVDQTRSPR SPSEEKEEVQ VAQSDVNIRN VFGSSDDEDA 180
181 EEYVRNDVEQ DEHRSPIEDE EGSEKDLRPD DMVLDDIIPE EDPQYESEAE HVEARYRERP 240
241 VGPPLEVEVP FRPPPGDPVK MNMIKVSNIM GIDPKPFDAK TFVEEDTFMT DEPGAKNRIR 300
301 LDNNIVRHRF VKSRDGKTYS ESNARFVRWS DGSLQLLIGN EVLNITEQDA KEDQNHLFIK 360
361 HEKGILQSQG RILKKMRFTP SSLTSNSHRL LTAIVESRQK KAFKVKNCVT DIDPEREKEK 420
421 REKAESQNLK ASTKLSQARE KIKRKYPLPV ERRQLSTGYL EDALDEDDED YRSNRGYEED 480
481 LEAEAQRERR ILNAKKSHKG IPGRSSMTSA RPSRRQMEYS ESEREESEYE TEEEEEEKSP 540
541 ARGRGKDSED EYEEDAEEDE EERGKSNRYS DEDEEEEEVA GGRAEKDHRG SGRKRKGIES 600
601 DEEESPPRKA PTHRRKAVID DSDED |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
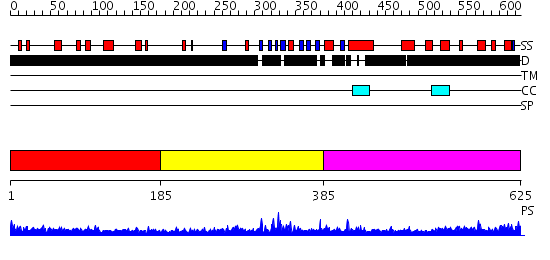
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..184] | 8.113997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [185..384] | 1.10097 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [385..625] | 1.083989 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)