













| General Information: |
|
| Name(s) found: |
gi|31711718
[NCBI NR]
gi|25405483 [NCBI NR] gi|15217969 [NCBI NR] gi|12325367 [NCBI NR] gi|12321677 [NCBI NR] gi|110735740 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 621 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: |
nucleic acid binding
[ISS]
RNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEFSTSKRPA TTATAAESVH FRLLCPATRT GAIIGKGGSV IRHLQSVTGS KIRVIDDIPV 60
61 PSEERVVLII APSGKKKDES NVCDSENPGS EEPKQEKGSE CAGTSGGDDE EAPSSAQMAL 120
121 LRVFERIVFG DDAATVDGDE LDKGESEGLC RMIVRGNQVD YLMSKGGKMI QKIREDSGAI 180
181 VRISSTDQIP PCAFPGDVVI QMNGKFSSVK KALLLVTNCL QESGAPPTWD ECPFPQPGYP 240
241 PEYHSMEYHP QWDHPPPNPM PEDVGPFNRP VVEEEVAFRL LCPADKVGSL IGKGGAVVRA 300
301 LQNESGASIK VSDPTHDSEE RIIVISAREN LERRHSLAQD GVMRVHNRIV EIGFEPSAAV 360
361 VARLLVHSPY IGRLLGKGGH LISEMRRATG ASIRVFAKDQ ATKYESQHDE IVQVIGNLKT 420
421 VQDALFQILC RLREAMFPGR LPFQGMGGPP PPFMGPYPEP PPPFGPRQYP ASPDRYHSPV 480
481 GPFHERHCHG PGFDRPPGPG FDRPPSPMSW TPQPGIDGHP GGMVPPDVNH GFALRNEPIG 540
541 SENPVMTSAN VEIVIPQAYL GHVYGENCSN LNYIKQVSGA NVVVHDPKAG TTEGLVVVSG 600
601 TSDQAHFAQS LLHAFILCGQ S |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
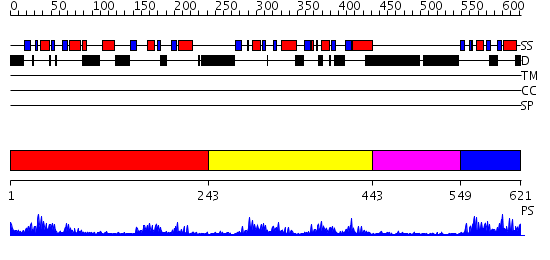
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..242] | 25.69897 | No description for 2annA was found. | |
| 2 | View Details | [243..442] | 22.69897 | Far upstream binding element, FBP, KH3 and KH4 domains | |
| 3 | View Details | [443..548] | 1.12 | No description for 2j63A was found. | |
| 4 | View Details | [549..621] | 17.09691 | HnRNP K, KH3 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 | No functions predicted. | ||||||||||||
| 2 |
|
||||||||||||
| 3 | No functions predicted. | ||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)