













| General Information: |
|
| Name(s) found: |
PFPA1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 614 amino acids |
Gene Ontology: |
|
| Cellular Component: |
pyrophosphate-dependent phosphofructokinase complex, alpha-subunit complex
[ISS]
|
| Biological Process: |
response to glucose stimulus
[IEP]
response to fructose stimulus [IEP] photosynthesis [IMP] response to sucrose stimulus [IEP] |
| Molecular Function: |
diphosphate-fructose-6-phosphate 1-phosphotransferase activity
[IMP][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSDFGIPRE LSPLQQLRSQ YKPELPPCLQ GTTVRVELGD GTTVAEAADS HTMARAFPHT 60
61 LGQPLAHFLR ETAQVPDAHI ITELPSVRVG IVFCGRQAPG GHNVIWGLFE ALKVHNAKST 120
121 LLGFLGGSEG LFAQKTLEIT DDILQTYKNQ GGYDLLGRTK DQIKTTEQVN AALKACTDLK 180
181 LDGLVIIGGV ISNTDAAHLA EFFAAAKCST KVVGVPVTTN GDLKNQFVEA NVGFDTICKV 240
241 NSQLISNACT DALSAEKYYY FIRLMGRKHS HVALECTLQS HPNMVILGEE VAASKLTIFD 300
301 IAKQICDAVQ ARAVEDKNHG VILIPEGLIV SIPEVYALLK EIHGLLRQGV SADKISTQLS 360
361 PWSSALFEFL PPFIKKQLLL HPESDDSAQL SQIETEKLLA YLVETEMNKR LKEGTYKGKK 420
421 FNAICHFFGY QARGSLPSKF DCDYAYVLGH ICYHILAAGL NGYMATVTNL KSPVNKWKCG 480
481 ATPITAMMTV KHWSQDASYT LTSIGRPAIH PAMVDLKGKA YDLLRQNAQK FLMEDLYRNP 540
541 GPLQYDGPGA DAKAVSLCVE DQDYMGRIKK LQEYLDQVRT IVKPGCSQDV LKAALSVMAS 600
601 VTDVLTTISS NGGQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
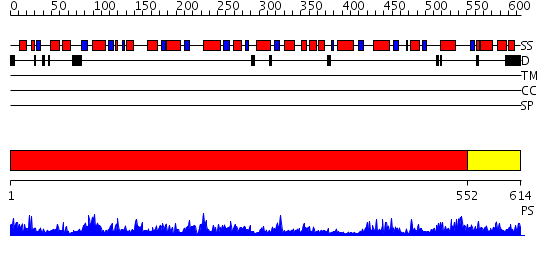
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..551] | 139.0 | Pyrophosphate-dependent phosphofructokinase | |
| 2 | View Details | [552..614] | 1.019986 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)