













| General Information: |
|
| Name(s) found: |
HMGCL_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 468 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA]
|
| Biological Process: |
metabolic process
[IEA]
leucine metabolic process [ISS] |
| Molecular Function: |
catalytic activity
[IEA]
hydroxymethylglutaryl-CoA lyase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQWNGVRRAH SIWCKRLTNN THLHHPSIPV SHFFTMSSLE EPLSFDKLPS MSTMDRIQRF 60
61 SSGACRPRDD VGMGHRWIEG RDCTTSNSCI DDDKSFAKES FPWRRHTRKL SEGEHMFRNI 120
121 SFAGRTSTVS GTLRESKSFK EQKYSTFSNE NGTSHISNKI SKGIPKFVKI VEVGPRDGLQ 180
181 NEKNIVPTSV KVELIQRLVS SGLPVVEATS FVSPKWVPQL ADAKDVMDAV NTLDGARLPV 240
241 LTPNLKGFQA AVSAGAKEVA IFASASESFS LSNINCTIEE SLLRYRVVAT AAKEHSVPVR 300
301 GYVSCVVGCP VEGPVLPSKV AYVVKELYDM GCFEISLGDT IGIGTPGSVV PMLEAVMAVV 360
361 PADKLAVHFH DTYGQALANI LVSLQMGISI VDSSIAGLGG CPYAKGASGN VATEDVVYML 420
421 NGLGVHTNVD LGKLIAAGDF ISKHLGRPNG SKAAVALNRR ITADASKI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
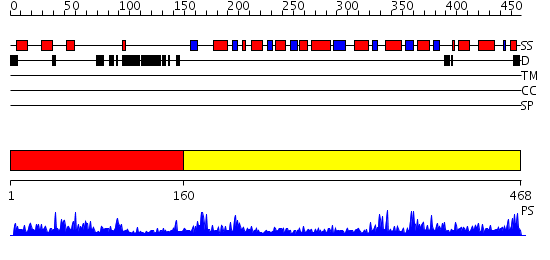
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..159] | 1.071981 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [160..468] | 92.69897 | Crystal Structure of Human HMG-CoA Lyase: Insights into Catalysis and the Molecular Basis for Hydroxymethylglutaric Aciduria |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)