













| General Information: |
|
| Name(s) found: |
SPP1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 423 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
sucrose biosynthetic process
[IEA][ISS]
metabolic process [IEA] |
| Molecular Function: |
magnesium ion binding
[IEA]
catalytic activity [IEA] sucrose-phosphatase activity [IEA][ISS] phosphatase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MERLTSPPRL MIVSDLDETM VDHHKDPENL ALLRFNSLWE DAYRHDSLLV FSTGRAQTMY 60
61 KKLRKEKPLL TPDVIITSVG TEIAYGNSMV PDDNWVEILN KKWDRGIVQE ETSKFPELTL 120
121 QGETEQRPHK LSFNIDKSKV KAVTKELSPR LEKRGVDVKF IFSGGNAFDV LAKGGGKGQA 180
181 LAYLLKKLKT EGKLPINTLA CGDSGNDTEL FTIPNVYGVM VSNAQEELLE WYAENAKDNA 240
241 NIIHASERCA GGITQAIGHF KLGPNLSPRD VSDFLECKAD NVNPGHEVVK FFLFYERWRR 300
301 GEVENCTTYT SSLKASCHPS GVFVHPSGAE KSLRDTIDEL GKYYGDKKGK KFRVWTDQVL 360
361 ATDTTPGTWI VKLDKWEQTG DERKCCTTTV KFTSKEGEGF VWEHVQQIWS EETEIKDDSN 420
421 WII |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
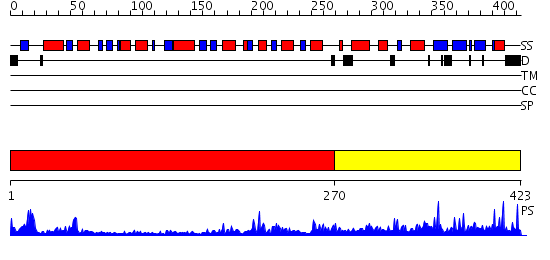
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..269] | 36.39794 | X-Ray structure of the sucrose-phosphatase (SPP) from Synechocystis sp. PCC6803 at 1.40 A resolution | |
| 2 | View Details | [270..423] | 5.49 | Structural Genomics, 1.5A crystal structure of a hypothetical protein PA1314 from Pseudomonas aeruginosa |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)