













| General Information: |
|
| Name(s) found: |
PMI1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 843 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
chloroplast relocation
[IMP]
[NOT]actin cytoskeleton organization [IMP] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGEYSGSRS SNTQLLAELE ALSENLYQKP QVSVGNRRTN SLALPRSSVP SLVTSADEVS 60
61 TARAEDLTVS KPRARRLSLS PWRSRPKLEV EEEENVTQSN RIVKKPEESS SGSGVKEEKK 120
121 GIWNWKPIRG LVRIGMQKLS CLLSVEVVAA QNLPASMNGL RLGVCVRKKE TKDGAVQTMP 180
181 CRVSQGSADF EETLFIKCHV YYSPANGKGS PAKFEARPFL FYLFAVDAKE LEFGRHVVDL 240
241 SELIQESVEK MNYEGARVRQ WDMNWGLSGK AKGGELALKL GFQIMEKDGG AGIYSKQGEF 300
301 GMKPSSKPKN FANSFGRKQS KTSFSVPSPK MTSRSEAWTP ASGVESVSDF HGMEHLNLDE 360
361 PEEKPEEKPV QKNDKPEQRA EDDQEEPDFE VVDKGVEFDD DLETEKSDGT IGERSVEMKE 420
421 QHVNVDDPRH IMRLTELDSI AKQIKALESM MKDESDGGDG ETESQRLDEE EQTVTKEFLQ 480
481 LLEDEETEKL KFYQHKMDIS ELRSGESVDD ESENYLSDLG KGIGCVVQTR DGGYLVSMNP 540
541 FDTVVMRKDT PKLVMQISKQ IVVLPEAGPA TGFELFHRMA GSGEELESKI SSLMAIDELM 600
601 GKTGEQVAFE GIASAIIQGR NKERANTSAA RTVAAVKTMA NAMSSGRRER IMTGIWNVEE 660
661 NPLTSAEEVL AVSLQKLEEM VVEGLKIQAD MVDDEAPFEV SAAKGQKNPL ESTIPLEEWQ 720
721 KEHRTQQKLT VLATVQLRDP TRRYEAVGGT VVVAVQAEEE EEKGLKVGSL HIGGVKKDAA 780
781 EKRRLTAAQW LVEHGMGKKG KKKSNIKKKE KEEEEEEMLW SLSSRVMADM WLKSIRNPDV 840
841 KLH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
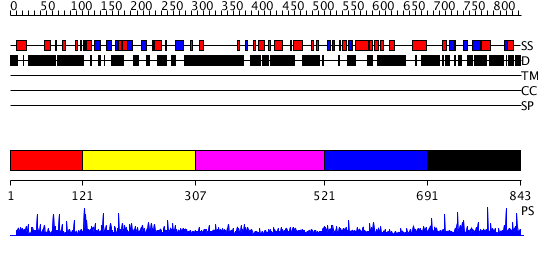
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..120] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [121..306] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [307..520] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [521..690] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [691..843] | 1.009985 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)