













| General Information: |
|
| Name(s) found: |
GGAP2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 431 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
response to ozone
[IEP]
response to jasmonic acid stimulus [IEP] L-ascorbic acid biosynthetic process [IMP] |
| Molecular Function: |
quercetin 4'-O-glucosyltransferase activity
[IDA]
GDP-D-glucose phosphorylase activity [IDA] galactose-1-phosphate guanylyltransferase (GDP) activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLLKIKRVPT VVSNYQKDET VEEGGCGRNC LSKCCINGAR LPLYTCKNLD KSVGENTESP 60
61 VTFLESLVIG EWEDRFQRGL FRYDVTACET KVIPGKYGFI AQLNEGRHLK KRPTEFRVDK 120
121 VLQPFDGNKF NFTKVGQEEL LFQFKASTND DDSEIQFLAS MPLDADNSPS VVAINVSPIE 180
181 YGHVLLIPRV LDCLPQRIDH KSLLLALQMA AEADNPYFRL GYNSLGAFAT INHLHFQAYY 240
241 LAMQFPIEKA SSLKITTTNN GVKISKLLNY PVRGLLVEGG NTIKDLADTV SDASVCLQNN 300
301 NIPFNILISD SGKRIFLLPQ CYAEKQALGE VSSTLLDTQV NPAVWEMSGH MVLKRKEDYE 360
361 GASEEKAWRL LAEVSLSEER FREVNTMIFD AIGFSSHEEE EEEELEEQNS MNGGSFTIVH 420
421 CPSVKEEAVS N |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
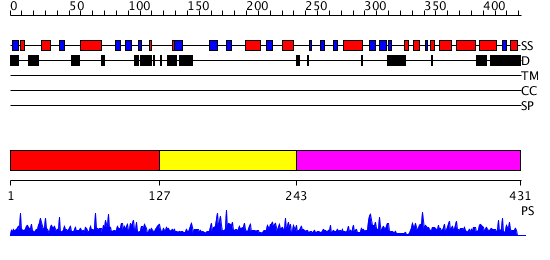
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..126] | 3.072985 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [127..242] | 2.65 | Crystal structure of rabbit Hint complexed with N-ethylsulfamoyladenosine | |
| 3 | View Details | [243..431] | 2.080982 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)