













| General Information: |
|
| Name(s) found: |
UPL4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1502 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ubiquitin ligase complex
[ISS]
|
| Biological Process: |
protein ubiquitination
[ISS]
|
| Molecular Function: |
ubiquitin-protein ligase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MENRGQKRME VVEELPADKR ACNSQDFRPS TSGSSVQAQA NDTNPGHENV DADMDTSSSA 60
61 SPSSRSDEEE QEEQDKEDSD YGSCDSDEED PRQRVLQDYQ RQRSSGDHGK LKSLLLNLTG 120
121 ETDPSGQLSR LTELCEVLSF STEESLSSVM ANMLSPVLVK LAKHENNADI MLLAIRAITY 180
181 LCDVYPPSVE FLVRHDTIPA LCQRLLTIEY LDVAEQCLQA LEKISRDEPV ACLNAGAIMA 240
241 VLSFIDFFST SIQRVAISTV VNICKQLSSE SPSPFMDAVP ILCTLLQYED RQLVENVAIC 300
301 LTKIADQASE SPAMLDQLCR HGLINESTHL LNLNSRTTLS QPVYNGVIGM LRKLSSGSAL 360
361 AFRTLYELNI GYSLKEIMST YDISHSVSST HPINACSNQV HEVLKLVIEL LPASPVEDNQ 420
421 LASEKESFLV NQPDLLQQFG RDMLPVMIQV LNSGANVYVS YGCLSAIHKL TCLSKSGDIV 480
481 ELLKNTNMSS VLAGILSRKD HHVIVVALQV AEVLLEKYRD TFLNSFIKEG VFFAIEALLS 540
541 SDRGQQNQGS ADLSQKPVTK EIVKCLCQSF ERSLSSSSQT CKIEKDSVYV LATRIKEGFF 600
601 GPEVFNSEKG LTDVLQNLKN LSVALSELMT VPIDAHVLHD EKFFSIWNQI MERLNGRESV 660
661 STFEFIESGV VKSLASYLSN GLYQRKLSKG GPECDSLPFI GKRFEVFTRL LWSDGEATSS 720
721 LLIQKLQNSL SSLENFPIVL SQFLKQKNSF AAIPNGRCTS YPCLKVRFLK AEGETSLRDY 780
781 SQDFVTVDPL CYLDAVDQYL WPKVNIEPID SVEAKDQAIE CQSSQLQSTS ISCQAESSSP 840
841 MEIDSESSDA SQLQGSQVED QTQLPGQQNA SSSETSSEKE DAVPRLLFRL EGLELDRSLT 900
901 VYQAILLHKL KSESEATNDS KLSGPHNITY ERSAQLGDSR ENLFPPGSME DDEYRPFLSY 960
961 LFTHRLALRL KGSSHPPYDI LFLLKSLEGM NRFLFHLISL ERINAFGEGR LENLDDLRVQ1020
1021 VRPVPHSEFV SSKLTEKLEQ QLRDSFAVST CGLPPWFNDL MDSCPCLFSF EAKSKYFRLA1080
1081 AFGSQKIRHH PQHLSSSNVH GEARPVTGSL PRKKFLACRE NILESAAKMM ELYGNQKVVI1140
1141 EVEYSEEVGT GLGPTLEFYT LVSRAFQNPD LGMWRNDCSF IVGKPVEHSG VLASSSGLFP1200
1201 RPWSGTSTTS DVLQKFVLLG TVVAKALQDG RVLDLPLSKA FYKLILGQEL SSFDIHFVDP1260
1261 ELCKTLVELQ ALVRRKKLFA EAHGDSGAAK CDLSFHGTKI EDLCLEFALP GYTDYDLAPY1320
1321 SANDMVNLDN LEEYIKGIVN ATVCNGIQKQ VEAFRSGFNQ VFSIEHLRIF NEEELETMLC1380
1381 GECDLFSMNE VLDHIKFDHG YTSSSPPVEY LLQILHEFDR EQQRAFLQFV TGSPRLPHGG1440
1441 LASLSPKLTI VRKHGSDSSD TDLPSVMTCA NYLKLPPYSS KEKMKEKLIY AITEGQGSFH1500
1501 LS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
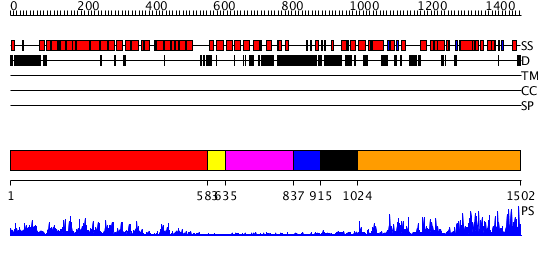
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..582] | 98.221849 | Crystal structure of the exportin Cse1p complexed with its cargo ( Kap60p) and RanGTP | |
| 2 | View Details | [583..634] | 19.0 | AP1 clathrin adaptor core | |
| 3 | View Details | [635..836] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [837..914] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [915..1023] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1024..1502] | 110.0 | Regulation of Smurf2 Ubiquitin Ligase Activity by Anchoring the E2 to the HECT domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.78 |
Source: Reynolds et al. (2008)