













| General Information: |
|
| Name(s) found: |
DUS3L_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 700 amino acids |
Gene Ontology: |
|
| Cellular Component: |
vacuole
[IDA]
|
| Biological Process: |
tRNA processing
[IEA]
metabolic process [IEA] oxidation reduction [IEA] |
| Molecular Function: |
tRNA dihydrouridine synthase activity
[IEA]
FAD binding [IEA] catalytic activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDVIAEHAV DTSVEGDAKA ALPVQSTKQF SVYEATSEEL IERSMAPIKK EFLCPPPPSR 60
61 SVKQNDAADV RAPQSGLVQE KKSKRQLKRE RREVLIRITS VLQSTINLCP QVARTEDVDS 120
121 CQYKDKCRFN HDIEAFKAQK ADDIEGQCPF VASGMKCAYG LSCRFLGSHR DITGNSDDKE 180
181 KSEMNFFNKE TQRLLWKNKM TFTNADAKLK SLGLLGHAKK SNAAEEITAE KTQNGMNGTQ 240
241 ATEVAVDSAV SSEHTSEMIQ DVDIPGPLET EEVRPMKKAK SEDQKNSKTG DVGGVYDGVK 300
301 LEEETKKNGY PTSKANVEDE DSIKIVETDS SLKLHPREKK KLIDFRDKLY LAPLTTVGNL 360
361 PFRRLCKVLG ADVTCGEMAM CTNLLQGQAS EWALLRRHSS EDLFGVQICG SYPDTVSRVV 420
421 ELIDRECTVD FIDINMGCPI DMVVNKSAGS ALLNKPLRMK NIVEVSSSIV ETPITIKVRT 480
481 AFFEGKNRID SLIADIGNWG ATAVTIHGRS RQQRYSKSAD WDYIYQCTKN ATTNLQVIGN 540
541 GDVYSYLDWN KHKSDCPELS SCMIARGALI KPWIFTEIKE QRHWDITSGE RLNIMKDFVR 600
601 FGLQHWGSDT KGVETTRHFL LEWLSYTFRY IPVGLLDVIP QQINWRPPSY FGRDDLETLM 660
661 MSESAGDWVR ISEMLLGKVP EGFTFAPKHK SNAYDRAENG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
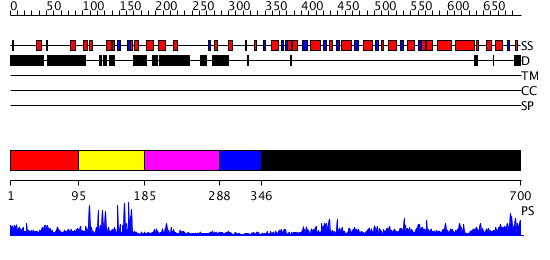
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..94] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [95..184] | 18.69897 | Structural Basis for Recognition of the mRNA Class II AU-Rich Element by the Tandem Zinc Finger Domain of TIS11d | |
| 3 | View Details | [185..287] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [288..345] | 18.154902 | Trimethylamine dehydrogenase, N-terminal domain; Trimethylamine dehydrogenase, C-terminal domain; Trimethylamine dehydrogenase, middle domain | |
| 5 | View Details | [346..700] | 50.522879 | Crystal structure of a putative flavin oxidoreductase with flavin |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)