













| General Information: |
|
| Name(s) found: |
YODA_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 883 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
embryonic development ending in seed dormancy
[IMP]
stomatal complex morphogenesis [IMP] |
| Molecular Function: |
ATP binding
[IEA]
protein kinase activity [IEA] protein serine/threonine kinase activity [IEA] kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPWWSKSKDE KKKTNKESII DAFNRKLGFA SEDRSSGRSR KSRRRRDEIV SERGAISRLP 60
61 SRSPSPSTRV SRCQSFAERS PAVPLPRPIV RPHVTSTDSG MNGSQRPGLD ANLKPSWLPL 120
121 PKPHGATSIP DNTGAEPDFA TASVSSGSSV GDIPSDSLLS PLASDCENGN RTPVNISSRD 180
181 QSMHSNKNSA EMFKPVPNKN RILSASPRRR PLGTHVKNLQ IPQRDLVLCS APDSLLSSPS 240
241 RSPMRSFIPD QVSNHGLLIS KPYSDVSLLG SGQCSSPGSG YNSGNNSIGG DMATQLFWPQ 300
301 SRCSPECSPV PSPRMTSPGP SSRIQSGAVT PLHPRAGGST TGSPTRRLDD NRQQSHRLPL 360
361 PPLLISNTCP FSPTYSAATS PSVPRSPARA EATVSPGSRW KKGRLLGMGS FGHVYLGFNS 420
421 ESGEMCAMKE VTLCSDDPKS RESAQQLGQE ISVLSRLRHQ NIVQYYGSET VDDKLYIYLE 480
481 YVSGGSIYKL LQEYGQFGEN AIRNYTQQIL SGLAYLHAKN TVHRDIKGAN ILVDPHGRVK 540
541 VADFGMAKHI TAQSGPLSFK GSPYWMAPEV IKNSNGSNLA VDIWSLGCTV LEMATTKPPW 600
601 SQYEGVPAMF KIGNSKELPD IPDHLSEEGK DFVRKCLQRN PANRPTAAQL LDHAFVRNVM 660
661 PMERPIVSGE PAEAMNVASS TMRSLDIGHA RSLPCLDSED ATNYQQKGLK HGSGFSISQS 720
721 PRNMSCPISP VGSPIFHSHS PHISGRRSPS PISSPHALSG SSTPLTGCGG AIPFHHQRQT 780
781 TVNFLHEGIG SSRSPGSGGN FYTNSFFQEP SRQQDRSRSS PRTPPHVFWD NNGSIQPGYN 840
841 WNKDNQPVLS DHVSQQLLSE HLKLKSLDLR PGFSTPGSTN RGP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
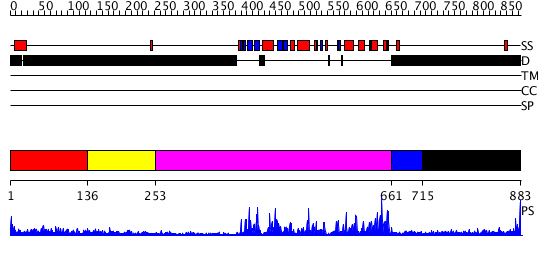
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..135] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [136..252] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [253..660] | 58.30103 | No description for 2j0jA was found. | |
| 4 | View Details | [661..714] | 89.69897 | No description for 2qnjA was found. | |
| 5 | View Details | [715..883] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)