













| General Information: |
|
| Name(s) found: |
EMF1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1096 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
meristem determinacy
[IMP]
shoot development [IGI][IMP] histone methylation [IMP] negative regulation of flower development [IMP] |
| Molecular Function: |
transcription regulator activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSSIKINSI SIDLAGAANE IDMVKCDHFS MRGFVAETRE RDLRKCWPFS EESVSLVDQQ 60
61 SYTLPTLSVP KFRWWHCMSC IKDIDAHGPK DCGLHSNSKA IGNSSVIESK SKFNSLTIID 120
121 HEKEKKTDIA DNAIEEKVGV NCENDDQTAT TFLKKARGRP MGASNVRSKS RKLVSPEQVG 180
181 NNRSKEKLNK PSMDISSWKE KQNVDQAVTT FGSSEIAGVV EDTPPKATKN HKGIRGLMEC 240
241 DNGSSESINL AMSGLQRRKS RKVRLLSELL GNTKTSGGSN IRKEESALKK ESVRGRKRKL 300
301 LPENNYVSRI LSTMGATSEN ASKSCDSDQG NSESTDSGFD RTPFKGKQRN RRFQVVDEFV 360
361 PSLPCETSQE GIKEHDADPS KRSTPAHSLF TGNDSVPCPP GTQRTERKLS LPKKKTKKPV 420
421 IDNGKSTVIS FSNGIDGSQV NSHTGPSMNT VSQTRDLLNG KRVGGLFDNR LASDGYFRKY 480
481 LSQVNDKPIT SLHLQDNDYV RSRDAEPNCL RDFSSSSKSS SGGWLRTGVD IVDFRNNNHN 540
541 TNRSSFSNLK LRYPPSSTEV ADLSRVLQKD ASGADRKGKT VMVQEHHGAP RSQSHDRKET 600
601 TTEEQNNDDI PMEIVELMAK NQYERCLPDK EEDVSNKQPS QETAHKSKNA LLIDLNETYD 660
661 NGISLEDNNT SRPPKPCSSN ARREEHFPMG RQQNSHDFFP ISQPYVPSPF GIFPPTQENR 720
721 ASSIRFSGHN CQWLGNLPTV GNQNPSPSSF RVLRACDTCQ SVPNQYREAS HPIWPSSMIP 780
781 PQSQYKPVSL NINQSTNPGT LSQASNNENT WNLNFVAANG KQKCGPNPEF SFGCKHAAGV 840
841 SSSSSRPIDN FSSESSIPAL HLLSLLDPRL RSTTPADQHG NTKFTKRHFP PANQSKEFIE 900
901 LQTGDSSKSA YSTKQIPFDL YSKRFTQEPS RKSFPITPPI GTSSLSFQNA SWSPHHQEKK 960
961 TKRKDTFAPV YNTHEKPVFA SSNDQAKFQL LGASNSMMLP LKFHMTDKEK KQKRKAESCN1020
1021 NNASAGPVKN SSGPIVCSVN RNPADFTIPE PGNVYMLTGE HLKVRKRTTF KKKPAVCKQD1080
1081 AMKQTKKPVC PPTQNA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
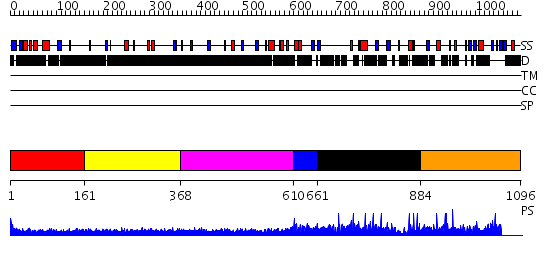
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..160] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [161..367] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [368..609] | 2.298994 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [610..660] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [661..883] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [884..1096] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)