













| General Information: |
|
| Name(s) found: |
SMC5_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1053 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS]
chromosome [IEA] |
| Biological Process: |
sister chromatid cohesion
[IMP]
chromosome segregation [ISS] |
| Molecular Function: |
ATP binding
[IEA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSERRAKRPK ISRGEDDFLP GNIIEIELHN FMTFNHLVCK PGSRLNLVIG PNGSGKSSLV 60
61 CAIALCLGGE PQLLGRATSV GAYVKRGEDS GYVKISLRGN TREENLTIFR KIDTRNKSEW 120
121 MFNGSTVSKK DIVEIIQKFN IQVNNLTQFL PQDRVCEFAK LTPVQLLEET EKAVGDPQLP 180
181 VHHRALVEKS RDLKQLERAV AKNGETLNQL KALVDEQEKD VERVRQRELF LTKVDSMKKK 240
241 LPWLKYDMKK AEYMDAKKRM KEAEKKLDEA AKNLNSMKEP IEKQKKEKAE TDSKCKKVKN 300
301 LMDANGRNRC HLLEKEDEAD ARVVATYKEL EELKKQEEHR QERILKATED LVAAERELQN 360
361 LPVYERPVAK LEELSSQVTE LHHSINGKKN QKEDNEKLLS QKRYTLRQCV DKLKDMENAN 420
421 NKLLKALANS GADRIFDAYQ WVQQNRHEFK REVYGPVLVE VNVPNRENAC FLEGHVSFYI 480
481 WKSFITQDPE DRDLLVKNLK RFDVPVLNYV GNSGNQKAPF HISDQMRSLG IHARLDQIFD 540
541 APDAVKEVLN SQFGLEDSYI GSKITDQRAE EVYKLGIKDF WTPDNHYRWS SSRYGGHSSA 600
601 SVDSVYQSRL LLCGVDVGEL EKLRSRKEEL EDSILFMEET HKSLQTEQRR LEEEAAKLHK 660
661 EREEIVNVSY LEKKKRRELE SRYQQRKTKL ESLEQEEDMD ASVAKLIDQA SRANADRYTY 720
721 AINLKKLLVE AVAHKWSYAE KHMASIELER KIRESEINIK QYEKTAQQLS LAVEYCKKEV 780
781 EGKQQRLATA KRDAESVATI TPELKKEFME MPTTVEELEA AIQDNLSQAN SILFINENIL 840
841 QEYEHRQSQI YTISTKLETD KRDLSICMKE IDSLKEKWLP TLRQLVGQIN ETFSHNFQEM 900
901 AVAGEVSLDE RDTDFDQYGI HIKVKFRESG QLQVLSSHHQ SGGERSVSTI LYLVSLQDLT 960
961 NCPFRVVDEI NQGMDPINER KMFQQLVRAA SQPNTPQCFL LTPKLLPELE YSEACSILNI1020
1021 MNGPYIAEPS KVWSLGDSWG SLNRRRTEAS QCS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
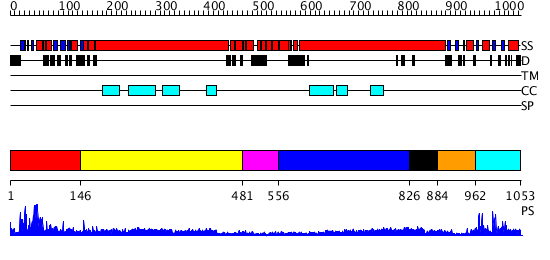
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..145] | 23.522879 | Sc Smc1hd:Scc1-C complex, ATPgS | |
| 2 | View Details | [146..480] | 21.0 | Heavy meromyosin subfragment | |
| 3 | View Details | [481..555] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [556..825] | 3.09691 | Interferon-induced guanylate-binding protein 1 (GBP1), C-terminal domain; Interferon-induced guanylate-binding protein 1 (GBP1), N-terminal domain | |
| 5 | View Details | [826..883] | 9.045757 | Tropomyosin | |
| 6 | View Details | [884..961] | 9.045757 | Tropomyosin | |
| 7 | View Details | [962..1053] | 21.30103 | Structural biochemistry of ATP-driven dimerization and DNA stimulated activation of SMC ATPases. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)