













| General Information: |
|
| Name(s) found: |
gi|7801660
[NCBI NR]
gi|15228453 [NCBI NR] gi|11357786 [NCBI NR] |
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1752 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKNKQDSSSP ETNIDSNPSK RTRFQSIETP QSQITHSTTA DTNATVPLTS AFSRVFKDIT 60
61 NLGYSSVGSF LTPSNNQQSY SAKKTHSTNI GSTVVSGLAS MVQANSRHMK QSQAVVFKDM 120
121 TNLPSMGLSS ATGKGKASQN ESTTSCMFFH KQTTLQTEKD RFPRKNLQND FENAFHSPIT 180
181 QGQSSATINN DFHCEMSDDE SMPDFNYDSS STNATDEDQH YYLSPTEDDT DDESTISIPV 240
241 ADPSSDICQN NANPTEPTTP HIAKCAEHEK FIATLKKTAA KAAANPNLPQ PKKRGRPRLN 300
301 KDTTNSAETA TKSAYLDHGD ATYKCNYCGA LMWFAERINK KQQNKSPTFT LCCGKGNVKL 360
361 PLLKDSPALI NNLLTGDDAL SRNFRENIRI YNMIFAMTSL GGRVDNSMPK GKGPNMFRLQ 420
421 GGNYHLIGSL KPNPGDYAKY SQLYIVDTEN EVDNRATVIN KGKGRRNTPA KQKLKKEVIE 480
481 ALIEMLNKVN PYVDKFRQAR ERIQDDNDEP FHMRIVADRK GVDRRTYSMP TSSEVAALIP 540
541 GGFQPSMFDR DIVLEEKTTG HLTRISQIHI SYLALQYPLI LCYGEDGYTP GIEKCLPNSA 600
601 KKKKKKCISM RQWFAFRIQE RPNECKTLTR SKRLFQQFLC DAYTTIESNR LSYIKFKQSK 660
661 LRCENYNSLK KASEAGTTSM NEEGNQVLIP SSLTGGPRYM VQNYYDAMAI CKHYGFPDLF 720
721 ITFTCNPKWP EITRHCQARG LSVDDRPDIV ARIFKIKLDS LMKDLTDGKM LGKTVASMHT 780
781 VEFQKRGLPH AHILLFMDAK SKLPTADDID KIISAEIPDK DKEPELYEVI KNSMIHGPCG 840
841 AANMNSPCMV EGKCSKQYPK KHQDITKVGK DGYPIYRRRM TEDYIEKGGF KCDNGYVVPY 900
901 NKKLSLRYQA HINVEWCNQS GSIKYLFKYI NKGADRVVFI VEPVNQDKTT ENATSGEPPN 960
961 STEKKKDEIK DWFDCRYVSA SEAVWRIYKF PLQDRSTAVQ RLSFHDEGKQ PVYAKPDADI1020
1021 EDVLERISNE DSMFMAWLTL NKNNDVGKNG KRARELLYSQ IPAYFTWDGK NKQWVKRIRG1080
1081 FSLGRINYVC RKMEVEYYLR VLLNIVKGPM SYDDIKTFNG VVYPSFKEAC FARGILDDDQ1140
1141 VYIDGLHEAS QFCFGDYLRN FFAMLLLSDS LARPEHVWSE TWHLLAEDIE NKKREDFKNP1200
1201 DLKLTLAEIR NYTLQEIEKI MLRNGATLKE IQDFPQPSRE GIDNSNRLVV DELRYNIDSN1260
1261 LKEKHDEWFQ MLNTEQRGIY DEITGAVFND LGGVFFIYGF GGTGKTFIWK TLAAAVRSRG1320
1321 QIVLNVASSG IASLLLEGGR TAHSRFAIPL NPDEFSVCKI TPKSDLANLI KEASLIIWDE1380
1381 APMMSKFCFE SLDKSFYDIL NNKDNKVFGG KVVVFGGDFR QVLPVINGAG RVEIVMSSLN1440
1441 ASYLWDHCKV LKLTKNMRLL SGGLSSEEAK EIQQFSDWLL AVGDGRINEP NDGEALIDIP1500
1501 EELLIKEAGN PIEAISKEIY GDPSELHMIN DPKFFQRRAI LAPTNEDVNT INQYMLEHLK1560
1561 SEERIYLSAD SIDPTDSDSL ANPVITPDFL NSIQLTGMPH HALRLKVGAP VMLLRNLDPK1620
1621 GGLCNGTRLQ ITQLAKQVVQ AKVITRDRIG DIVLIPLINL TPSDTKLPFK MRRRQFPLSV1680
1681 AFAMTINKSQ GQSLEQVGLY LPKPVFSHGQ LYVALSRVTS KKGLKILILD KDGNMQKQTT1740
1741 NVVFKEVFQN IV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
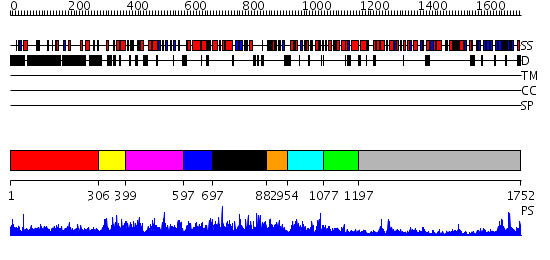
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..305] | 1.37 | THE BLUETONGUE VIRUS (BTV) CORE BINDS DSRNA | |
| 2 | View Details | [306..398] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [399..596] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [597..696] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [697..881] | 2.099961 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [882..953] | 1.098993 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [954..1076] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1077..1196] | 82.69897 | Crystal Structure Analysis of ClpB | |
| 9 | View Details | [1197..1752] | 30.154902 | RecBCD:DNA complex |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. | |||||||||
| 6 | No functions predicted. | |||||||||
| 7 | No functions predicted. | |||||||||
| 8 | No functions predicted. | |||||||||
| 9 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)