













| General Information: |
|
| Name(s) found: |
LOX4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 926 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IEA]
|
| Biological Process: |
jasmonic acid biosynthetic process
[TAS]
response to wounding [TAS] defense response [TAS] growth [TAS] |
| Molecular Function: |
oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen
[IEA]
metal ion binding [IEA] iron ion binding [IEA] electron carrier activity [IEA] lipoxygenase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALANEIMGS RLIFERSSSL ASPFHSRFSI KKKTQRTQFS INPFDPRPMR AVNSSGVVAA 60
61 ISEDLVKTLR ISTVGRKQEK EEEEEKSVKF KVRAVATVRN KNKEDFKETL VKHLDAFTDK 120
121 IGRNVVLELM STQVDPKTNE PKKSKAAVLK DWSKKSNSKA ERVHYTAEFT VDSAFGSPGA 180
181 ITVTNKHQKE FFLESITIEG FACGPVHFPC NSWVQSQKDH PSKRILFTNQ PYLPSETPSG 240
241 LRTLREKELE NLRGNGKGER KLSDRIYDYD VYNDIGNPDI SRELARPTLG GREFPYPRRC 300
301 RTGRSSTDTD MMSERRVEKP LPMYVPRDEQ FEESKQNTFA ACRLKAVLHN LIPSLKASIL 360
361 AEDFANFGEI DSLYKEGLLL KLGFQDDMFK KFPLPKIVTT LQKSSEGLLR YDTPKIVSKD 420
421 KYAWLRDDEF ARQAIAGINP VNIERVTSYP PVSNLDPEIY GPGLHSALTE DHIIGQLDGL 480
481 TVQQALETNR LFMVDYHDIY LPFLDRINAL DGRKAYATRT ILFLTRLGTL KPIAIELSLP 540
541 SQSSSNQKSK RVVTPPVDAT SNWMWQLAKA HVGSNDAGVH QLVNHWLRTH ACLEPFILAA 600
601 HRQLSAMHPI FKLLDPHMRY TLEINAVARQ TLISADGVIE SCFTAGQYGL EISSAAYKNK 660
661 WRFDMEGLPA DLIRRGMAVP DPTQPHGLKL LVEDYPYAND GLLLWSAIQT WVRTYVERYY 720
721 ANSNLIQTDT ELQAWYSESI NVGHADHRDA EWWPKLSTVE DLVSVITTII WLASAQHAAL 780
781 NFGQYPYGGY VPNRPPLMRR LIPDESDPEF TSFIEDPQKY FFSSMPSLLQ TTKFMAVVDT 840
841 LSTHSPDEEY IGERQQPSIW TGDAEIVDAF YGFSAEIGRI EKEIDKRNRD PSRRNRCGAG 900
901 VLPYELMAPS SEPGVTCRGV PNSVSI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
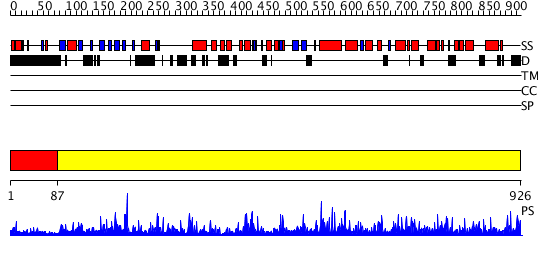
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..86] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [87..926] | 1000.0 | Lipoxigenase, C-terminal domain; Plant lipoxigenase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)